

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Squalene--hopene cyclase | ||
| Ligand | BDBM50128066 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_201950 (CHEMBL809137) | ||
| IC50 | 406.0±n/a nM | ||
| Citation |  Lenhart, A; Reinert, DJ; Aebi, JD; Dehmlow, H; Morand, OH; Schulz, GE Binding structures and potencies of oxidosqualene cyclase inhibitors with the homologous squalene-hopene cyclase. J Med Chem46:2083-92 (2003) [PubMed] Article Lenhart, A; Reinert, DJ; Aebi, JD; Dehmlow, H; Morand, OH; Schulz, GE Binding structures and potencies of oxidosqualene cyclase inhibitors with the homologous squalene-hopene cyclase. J Med Chem46:2083-92 (2003) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Squalene--hopene cyclase | |||
| Name: | Squalene--hopene cyclase | ||
| Synonyms: | SQHC_ALIAD | Squalene--hopene cyclase | Squalene-hopene cyclase | shc | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 71559.28 | ||
| Organism: | Alicyclobacillus acidocaldarius | ||
| Description: | ChEMBL_201951 | ||
| Residue: | 631 | ||
| Sequence: |
| ||
| BDBM50128066 | |||
| n/a | |||
| Name | BDBM50128066 | ||
| Synonyms: | CHEMBL66412 | METHYL-[4-(4-PIPERIDINE-1-YLMETHYL-PHENYL)-CYCLOHEXYL]-CARBAMINIC ACID-(4-CHLOROPHENYL)-ESTER | Methyl-[4-(4-piperidin-1-ylmethyl-phenyl)-cyclohexyl]-carbamic acid 4-chloro-phenyl ester | ||
| Type | Small organic molecule | ||
| Emp. Form. | C26H33ClN2O2 | ||
| Mol. Mass. | 441.005 | ||
| SMILES | CN([C@H]1CC[C@@H](CC1)c1ccc(CN2CCCCC2)cc1)C(=O)Oc1ccc(Cl)cc1 |wU:2.1,wD:5.8,(5.42,2.82,;5.42,1.28,;4.09,.51,;4.09,-1.03,;2.75,-1.8,;1.42,-1.03,;1.43,.51,;2.76,1.28,;.08,-1.78,;.09,-3.32,;-1.24,-4.09,;-2.58,-3.31,;-3.91,-4.08,;-5.25,-3.3,;-6.58,-4.08,;-7.91,-3.3,;-7.9,-1.76,;-6.57,-1,;-5.24,-1.77,;-2.58,-1.77,;-1.24,-1.01,;6.75,.51,;6.75,-1.03,;8.1,1.28,;9.43,.51,;9.43,-1.03,;10.76,-1.8,;12.09,-1.03,;13.42,-1.8,;12.09,.51,;10.76,1.28,)| | ||
| Structure | 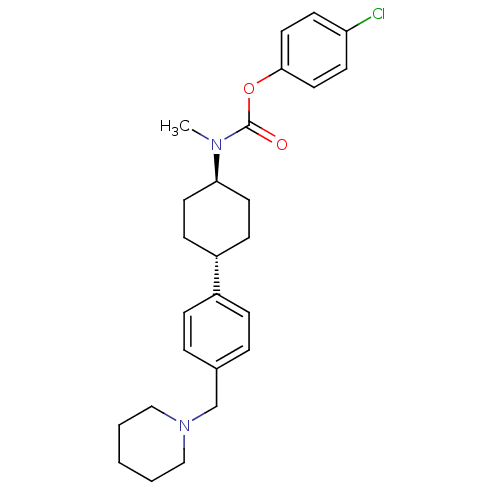
| ||