| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Squalene--hopene cyclase |
|---|
| Ligand | BDBM50128062 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_201950 (CHEMBL809137) |
|---|
| IC50 | 18±n/a nM |
|---|
| Citation |  Lenhart, A; Reinert, DJ; Aebi, JD; Dehmlow, H; Morand, OH; Schulz, GE Binding structures and potencies of oxidosqualene cyclase inhibitors with the homologous squalene-hopene cyclase. J Med Chem46:2083-92 (2003) [PubMed] Article Lenhart, A; Reinert, DJ; Aebi, JD; Dehmlow, H; Morand, OH; Schulz, GE Binding structures and potencies of oxidosqualene cyclase inhibitors with the homologous squalene-hopene cyclase. J Med Chem46:2083-92 (2003) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Squalene--hopene cyclase |
|---|
| Name: | Squalene--hopene cyclase |
|---|
| Synonyms: | SQHC_ALIAD | Squalene--hopene cyclase | Squalene-hopene cyclase | shc |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 71559.28 |
|---|
| Organism: | Alicyclobacillus acidocaldarius |
|---|
| Description: | ChEMBL_201951 |
|---|
| Residue: | 631 |
|---|
| Sequence: | MAEQLVEAPAYARTLDRAVEYLLSCQKDEGYWWGPLLSNVTMEAEYVLLCHILDRVDRDR
MEKIRRYLLHEQREDGTWALYPGGPPDLDTTIEAYVALKYIGMSRDEEPMQKALRFIQSQ
GGIESSRVFTRMWLALVGEYPWEKVPMVPPEIMFLGKRMPLNIYEFGSWARATVVALSIV
MSRQPVFPLPERARVPELYETDVPPRRRGAKGGGGWIFDALDRALHGYQKLSVHPFRRAA
EIRALDWLLERQAGDGSWGGIQPPWFYALIALKILDMTQHPAFIKGWEGLELYGVELDYG
GWMFQASISPVWDTGLAVLALRAAGLPADHDRLVKAGEWLLDRQITVPGDWAVKRPNLKP
GGFAFQFDNVYYPDVDDTAVVVWALNTLRLPDERRRRDAMTKGFRWIVGMQSSNGGWGAY
DVDNTSDLPNHIPFCDFGEVTDPPSEDVTAHVLECFGSFGYDDAWKVIRRAVEYLKREQK
PDGSWFGRWGVNYLYGTGAVVSALKAVGIDTREPYIQKALDWVEQHQNPDGGWGEDCRSY
EDPAYAGKGASTPSQTAWALMALIAGGRAESEAARRGVQYLVETQRPDGGWDEPYYTGTG
FPGDFYLGYTMYRHVFPTLALGRYKQAIERR
|
|
|
|---|
| BDBM50128062 |
|---|
| n/a |
|---|
| Name | BDBM50128062 |
|---|
| Synonyms: | (4-BROMOPHENYL)[4-({(2E)-4-[CYCLOPROPYL(METHYL)AMINO]BUT-2-ENYL}OXY)PHENYL]METHANONE | (4-Bromo-phenyl)-{4-[4-(cyclopropyl-methyl-amino)-but-2-enyloxy]-phenyl}-methanone | CHEMBL479653 | CHEMBL62855 | N-(4-(4-(4-bromobenzoyl)phenoxy)but-2-enyl)-N-methylcyclopropanaminium |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H22BrNO2 |
|---|
| Mol. Mass. | 400.309 |
|---|
| SMILES | CN(C\C=C\COc1ccc(cc1)C(=O)c1ccc(Br)cc1)C1CC1 |
|---|
| Structure | 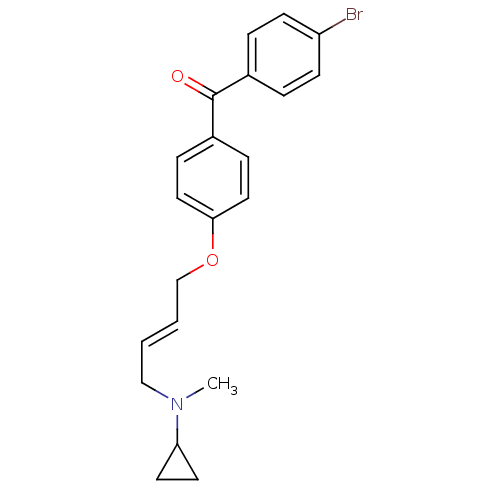
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lenhart, A; Reinert, DJ; Aebi, JD; Dehmlow, H; Morand, OH; Schulz, GE Binding structures and potencies of oxidosqualene cyclase inhibitors with the homologous squalene-hopene cyclase. J Med Chem46:2083-92 (2003) [PubMed] Article
Lenhart, A; Reinert, DJ; Aebi, JD; Dehmlow, H; Morand, OH; Schulz, GE Binding structures and potencies of oxidosqualene cyclase inhibitors with the homologous squalene-hopene cyclase. J Med Chem46:2083-92 (2003) [PubMed] Article