| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 1,3-beta-glucan synthase component GSC2 |
|---|
| Ligand | BDBM50572240 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2116620 (CHEMBL4825561) |
|---|
| IC50 | 220±n/a nM |
|---|
| Citation |  Eldhose, B; Pandrala, M; Xavier, C; Mohamed, AA; Srivastava, S; Sunkara, AD; Dobi, A; Malhotra, SV New Selective Inhibitors of ERG Positive Prostate Cancer: ERGi-USU-6 Salt Derivatives. ACS Med Chem Lett12:1703-1709 (2021) [PubMed] Article Eldhose, B; Pandrala, M; Xavier, C; Mohamed, AA; Srivastava, S; Sunkara, AD; Dobi, A; Malhotra, SV New Selective Inhibitors of ERG Positive Prostate Cancer: ERGi-USU-6 Salt Derivatives. ACS Med Chem Lett12:1703-1709 (2021) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 1,3-beta-glucan synthase component GSC2 |
|---|
| Name: | 1,3-beta-glucan synthase component GSC2 |
|---|
| Synonyms: | 1,3-beta-D-glucan-UDP glucosyltransferase | 1,3-beta-glucan synthase component GLS2 | FK506 sensitivity protein 2 | FKS2 | FKS2_YEAST | GLS2 | GSC2 | Glucan synthase of cerevisiae protein 2 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 216999.86 |
|---|
| Organism: | Saccharomyces cerevisiae |
|---|
| Description: | ChEMBL_11143 |
|---|
| Residue: | 1895 |
|---|
| Sequence: | MSYNDPNLNGQYYSNGDGTGDGNYPTYQVTQDQSAYDEYGQPIYTQNQLDDGYYDPNEQY
VDGTQFPQGQDPSQDQGPYNNDASYYNQPPNMMNPSSQDGENFSDFSSYGPPSGTYPNDQ
YTPSQMSYPDQDGSSGASTPYGNGVVNGNGQYYDPNAIEMALPNDPYPAWTADPQSPLPI
EQIEDIFIDLTNKFGFQRDSMRNMFDHFMTLLDSRSSRMSPEQALLSLHADYIGGDTANY
KKWYFAAQLDMDDEIGFRNMKLGKLSRKARKAKKKNKKAMQEASPEDTEETLNQIEGDNS
LEAADFRWKSKMNQLSPFEMVRQIALFLLCWGEANQVRFTPECLCFIYKCASDYLDSAQC
QQRPDPLPEGDFLNRVITPLYRFIRSQVYEIVDGRYVKSEKDHNKVIGYDDVNQLFWYPE
GIAKIVMEDGTRLIDLPAEERYLKLGEIPWDDVFFKTYKETRSWLHLVTNFNRIWIMHIS
VYWMYCAYNAPTFYTHNYQQLVDNQPLAAYKWATAALGGTVASLIQVAATLCEWSFVPRK
WAGAQHLSRRFWFLCVIMGINLGPVIFVFAYDKDTVYSTAAHVVGAVMFFVAVATLVFFS
VMPLGGLFTSYMKKSTRSYVASQTFTASFAPLHGLDRWMSYLVWVTVFAAKYAESYFFLI
LSLRDPIRILSTTSMRCTGEYWWGNKICKVQPKIVLGLMIATDFILFFLDTYLWYIVVNT
VFSVGKSFYLGISILTPWRNIFTRLPKRIYSKILATTDMEIKYKPKVLISQIWNAIIISM
YREHLLAIDHVQKLLYHQVPSEIEGKRTLRAPTFFVSQDDNNFETEFFPRDSEAERRISF
FAQSLSTPIPEPLPVDNMPTFTVLTPHYAERILLSLREIIREDDQFSRVTLLEYLKQLHP
VEWDCFVKDTKILAEETAAYENNEDEPEKEDALKSQIDDLPFYCIGFKSAAPEYTLRTRI
WASLRSQTLYRTISGFMNYSRAIKLLYRVENPEIVQMFGGNADGLERELEKMARRKFKFL
VSMQRLAKFKPHELENAEFLLRAYPDLQIAYLDEEPPLNEGEEPRIYSALIDGHCEILEN
GRRRPKFRVQLSGNPILGDGKSDNQNHALIFYRGEYIQLIDANQDNYLEECLKIRSVLAE
FEELGIEQIHPYTPGLKYEDQSTNHPVAIVGAREYIFSENSGVLGDVAAGKEQTFGTLFA
RTLAQIGGKLHYGHPDFINATFMTTRGGVSKAQKGLHLNEDIYAGMNAVLRGGRIKHCEY
YQCGKGRDLGFGTILNFTTKIGAGMGEQMLSREYYYLGTQLPIDRFLTFYYAHPGFHLNN
LFIQLSLQMFMLTLVNLHALAHESILCVYDRDKPITDVLYPIGCYNFHPAIDWVRRYTLS
IFIVFWIAFVPIVVQELIERGLWKATQRFFRHILSLSPMFEVFAGQIYSSALLSDIAVGG
ARYISTGRGFATSRIPFSILYSRFAGSAIYMGSRSMLMLLFGTVAHWQAPLLWFWASLSA
LIFAPFIFNPHQFAWEDFFLDYRDYIRWLSRGNNKYHRNSWIGYVRMSRSRVTGFKRKLV
GDESEKSAGDASRAHRTNLIMAEIIPCAIYAAGCFIAFTFINAQTGVKTTDEDRVNSTLR
IIICTLAPIVIDIGVLFFCMGLSCCSGPLLGMCCKKTGSVMAGIAHGIAVVVHIVFFIVM
WVLEGFSFVRMLIGVVTCIQCQRLIFHCMTVLLLTREFKNDHANTAFWTGKWYSTGLGYM
AWTQPTRELTAKVIELSEFAADFVLGHVILIFQLPVICIPKIDKFHSIMLFWLKPSRQIR
PPIYSLKQARLRKRMVRRYCSLYFLVLIIFAGCIVGPAVASAHVPKDLGSGLTGTFHNLV
QPRNVSNNDTGSQMSTYKSHYYTHTPSLKTWSTIK
|
|
|
|---|
| BDBM50572240 |
|---|
| n/a |
|---|
| Name | BDBM50572240 |
|---|
| Synonyms: | CHEMBL4857685 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H14N4O |
|---|
| Mol. Mass. | 242.2765 |
|---|
| SMILES | CN(C)c1ccc(\N=N\c2ccccn2)c(O)c1 |
|---|
| Structure | 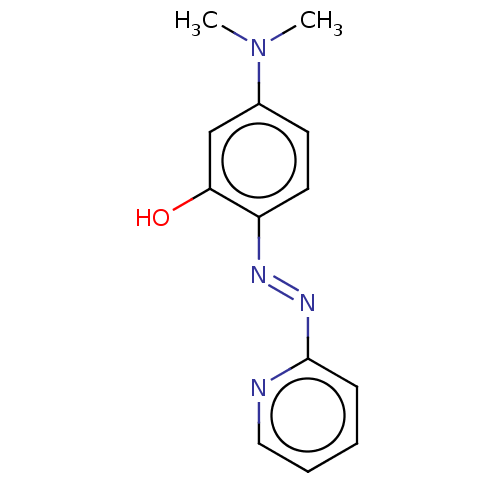
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Eldhose, B; Pandrala, M; Xavier, C; Mohamed, AA; Srivastava, S; Sunkara, AD; Dobi, A; Malhotra, SV New Selective Inhibitors of ERG Positive Prostate Cancer: ERGi-USU-6 Salt Derivatives. ACS Med Chem Lett12:1703-1709 (2021) [PubMed] Article
Eldhose, B; Pandrala, M; Xavier, C; Mohamed, AA; Srivastava, S; Sunkara, AD; Dobi, A; Malhotra, SV New Selective Inhibitors of ERG Positive Prostate Cancer: ERGi-USU-6 Salt Derivatives. ACS Med Chem Lett12:1703-1709 (2021) [PubMed] Article