| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Beta-secretase 2 |
|---|
| Ligand | BDBM50505569 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2147516 (CHEMBL5031862) |
|---|
| Ki | 0.120000±n/a nM |
|---|
| Citation |  Ueno, T; Matsuoka, E; Asada, N; Yamamoto, S; Kanegawa, N; Ito, M; Ito, H; Moechars, D; Rombouts, FJR; Gijsen, HJM; Kusakabe, KI Discovery of Extremely Selective Fused Pyridine-Derived ?-Site Amyloid Precursor Protein-Cleaving Enzyme (BACE1) Inhibitors with High In Vivo Efficacy through 10s Loop Interactions. J Med Chem64:14165-14174 (2021) [PubMed] Article Ueno, T; Matsuoka, E; Asada, N; Yamamoto, S; Kanegawa, N; Ito, M; Ito, H; Moechars, D; Rombouts, FJR; Gijsen, HJM; Kusakabe, KI Discovery of Extremely Selective Fused Pyridine-Derived ?-Site Amyloid Precursor Protein-Cleaving Enzyme (BACE1) Inhibitors with High In Vivo Efficacy through 10s Loop Interactions. J Med Chem64:14165-14174 (2021) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Beta-secretase 2 |
|---|
| Name: | Beta-secretase 2 |
|---|
| Synonyms: | AEPLC | ALP56 | ASP1 | ASP21 | Asp 1 | Aspartic-like protease 56 kDa | Aspartyl protease 1 | BACE2 | BACE2_HUMAN | Beta secretase 2 | Beta-secretase (BACE) | Beta-secretase 2 | Beta-secretase 2 (BACE-2) | Beta-secretase 2 precursor | Beta-site APP-cleaving enzyme 2 | Down region aspartic protease | Memapsin-1 | Membrane-associated aspartic protease 1 | beta-Secretase (BACE-2) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 56171.20 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9Y5Z0 |
|---|
| Residue: | 518 |
|---|
| Sequence: | MGALARALLLPLLAQWLLRAAPELAPAPFTLPLRVAAATNRVVAPTPGPGTPAERHADGL
ALALEPALASPAGAANFLAMVDNLQGDSGRGYYLEMLIGTPPQKLQILVDTGSSNFAVAG
TPHSYIDTYFDTERSSTYRSKGFDVTVKYTQGSWTGFVGEDLVTIPKGFNTSFLVNIATI
FESENFFLPGIKWNGILGLAYATLAKPSSSLETFFDSLVTQANIPNVFSMQMCGAGLPVA
GSGTNGGSLVLGGIEPSLYKGDIWYTPIKEEWYYQIEILKLEIGGQSLNLDCREYNADKA
IVDSGTTLLRLPQKVFDAVVEAVARASLIPEFSDGFWTGSQLACWTNSETPWSYFPKISI
YLRDENSSRSFRITILPQLYIQPMMGAGLNYECYRFGISPSTNALVIGATVMEGFYVIFD
RAQKRVGFAASPCAEIAGAAVSEISGPFSTEDVASNCVPAQSLSEPILWIVSYALMSVCG
AILLVLIVLLLLPFRCQRRPRDPEVVNDESSLVRHRWK
|
|
|
|---|
| BDBM50505569 |
|---|
| n/a |
|---|
| Name | BDBM50505569 |
|---|
| Synonyms: | CHEMBL4557670 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H18F3N5O2S |
|---|
| Mol. Mass. | 425.428 |
|---|
| SMILES | C[C@H]1SC(N)=N[C@@](C)([C@H]1F)c1cc(NC(=O)c2cnc(OCF)cn2)ccc1F |r,c:4| |
|---|
| Structure | 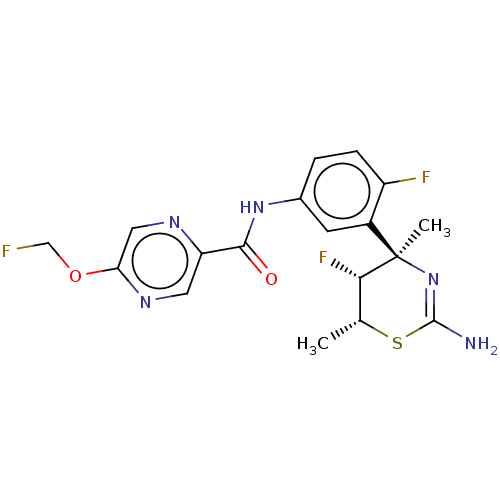
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ueno, T; Matsuoka, E; Asada, N; Yamamoto, S; Kanegawa, N; Ito, M; Ito, H; Moechars, D; Rombouts, FJR; Gijsen, HJM; Kusakabe, KI Discovery of Extremely Selective Fused Pyridine-Derived ?-Site Amyloid Precursor Protein-Cleaving Enzyme (BACE1) Inhibitors with High In Vivo Efficacy through 10s Loop Interactions. J Med Chem64:14165-14174 (2021) [PubMed] Article
Ueno, T; Matsuoka, E; Asada, N; Yamamoto, S; Kanegawa, N; Ito, M; Ito, H; Moechars, D; Rombouts, FJR; Gijsen, HJM; Kusakabe, KI Discovery of Extremely Selective Fused Pyridine-Derived ?-Site Amyloid Precursor Protein-Cleaving Enzyme (BACE1) Inhibitors with High In Vivo Efficacy through 10s Loop Interactions. J Med Chem64:14165-14174 (2021) [PubMed] Article