| Reaction Details |
|---|
 | Report a problem with these data |
| Target | GABA-A receptor; alpha-5/beta-3/gamma-2 |
|---|
| Ligand | BDBM50144862 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_70550 (CHEMBL680669) |
|---|
| Ki | 3±n/a nM |
|---|
| Citation |  Street, LJ; Sternfeld, F; Jelley, RA; Reeve, AJ; Carling, RW; Moore, KW; McKernan, RM; Sohal, B; Cook, S; Pike, A; Dawson, GR; Bromidge, FA; Wafford, KA; Seabrook, GR; Thompson, SA; Marshall, G; Pillai, GV; Castro, JL; Atack, JR; MacLeod, AM Synthesis and biological evaluation of 3-heterocyclyl-7,8,9,10-tetrahydro-(7,10-ethano)-1,2,4-triazolo[3,4-a]phthalazines and analogues as subtype-selective inverse agonists for the GABA(A)alpha5 benzodiazepine binding site. J Med Chem47:3642-57 (2004) [PubMed] Article Street, LJ; Sternfeld, F; Jelley, RA; Reeve, AJ; Carling, RW; Moore, KW; McKernan, RM; Sohal, B; Cook, S; Pike, A; Dawson, GR; Bromidge, FA; Wafford, KA; Seabrook, GR; Thompson, SA; Marshall, G; Pillai, GV; Castro, JL; Atack, JR; MacLeod, AM Synthesis and biological evaluation of 3-heterocyclyl-7,8,9,10-tetrahydro-(7,10-ethano)-1,2,4-triazolo[3,4-a]phthalazines and analogues as subtype-selective inverse agonists for the GABA(A)alpha5 benzodiazepine binding site. J Med Chem47:3642-57 (2004) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| GABA-A receptor; alpha-5/beta-3/gamma-2 |
|---|
| Name: | GABA-A receptor; alpha-5/beta-3/gamma-2 |
|---|
| Synonyms: | GABA receptor alpha-5/beta-3/gamma-2 subunit | Gamma-aminobutyric acid receptor subunit alpha-5/beta-3/gamma-2 |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of ChEMBL is 2148177 |
|---|
| Components: | This complex has 3 components. |
|---|
| Component 1 |
| Name: | Gamma-aminobutyric acid receptor subunit gamma-2 |
|---|
| Synonyms: | GABA(A) receptor subunit gamma-2 | GABRG2 | GBRG2_HUMAN |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 54172.74 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | EBI_217 |
|---|
| Residue: | 467 |
|---|
| Sequence: | MSSPNIWSTGSSVYSTPVFSQKMTVWILLLLSLYPGFTSQKSDDDYEDYASNKTWVLTPK
VPEGDVTVILNNLLEGYDNKLRPDIGVKPTLIHTDMYVNSIGPVNAINMEYTIDIFFAQT
WYDRRLKFNSTIKVLRLNSNMVGKIWIPDTFFRNSKKADAHWITTPNRMLRIWNDGRVLY
TLRLTIDAECQLQLHNFPMDEHSCPLEFSSYGYPREEIVYQWKRSSVEVGDTRSWRLYQF
SFVGLRNTTEVVKTTSGDYVVMSVYFDLSRRMGYFTIQTYIPCTLIVVLSWVSFWINKDA
VPARTSLGITTVLTMTTLSTIARKSLPKVSYVTAMDLFVSVCFIFVFSALVEYGTLHYFV
SNRKPSKDKDKKKKNPAPTIDIRPRSATIQMNNATHLQERDEEYGYECLDGKDCASFFCC
FEDCRTGAWRHGRIHIRIAKMDSYARIFFPTAFCLFNLVYWVSYLYL
|
|
|
|---|
| Component 2 |
| Name: | Gamma-aminobutyric acid receptor subunit beta-3 |
|---|
| Synonyms: | GABA A receptor alpha-4/beta-3/gamma-2 | GABA receptor beta-3 subunit | GABA-A receptor | GABRB3 | GBRB3_HUMAN | agonist GABA site |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 54130.51 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_448071 |
|---|
| Residue: | 473 |
|---|
| Sequence: | MWGLAGGRLFGIFSAPVLVAVVCCAQSVNDPGNMSFVKETVDKLLKGYDIRLRPDFGGPP
VCVGMNIDIASIDMVSEVNMDYTLTMYFQQYWRDKRLAYSGIPLNLTLDNRVADQLWVPD
TYFLNDKKSFVHGVTVKNRMIRLHPDGTVLYGLRITTTAACMMDLRRYPLDEQNCTLEIE
SYGYTTDDIEFYWRGGDKAVTGVERIELPQFSIVEHRLVSRNVVFATGAYPRLSLSFRLK
RNIGYFILQTYMPSILITILSWVSFWINYDASAARVALGITTVLTMTTINTHLRETLPKI
PYVKAIDMYLMGCFVFVFLALLEYAFVNYIFFGRGPQRQKKLAEKTAKAKNDRSKSESNR
VDAHGNILLTSLEVHNEMNEVSGGIGDTRNSAISFDNSGIQYRKQSMPREGHGRFLGDRS
LPHKKTHLRRRSSQLKIKIPDLTDVNAIDRWSRIVFPFTFSLFNLVYWLYYVN
|
|
|
|---|
| Component 3 |
| Name: | Gamma-aminobutyric acid receptor subunit alpha-5 |
|---|
| Synonyms: | GABA receptor alpha-5 subunit | GABA(A) receptor subunit alpha-5 (GABAAa5) | GABRA5 | GBRA5_HUMAN | Gamma-aminobutyric acid receptor subunit alpha-5 (GABA(A)) | Gamma-aminobutyric acid receptor subunit alpha-5 (GABAA) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52160.59 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P31644 |
|---|
| Residue: | 462 |
|---|
| Sequence: | MDNGMFSGFIMIKNLLLFCISMNLSSHFGFSQMPTSSVKDETNDNITIFTRILDGLLDGY
DNRLRPGLGERITQVRTDIYVTSFGPVSDTEMEYTIDVFFRQSWKDERLRFKGPMQRLPL
NNLLASKIWTPDTFFHNGKKSIAHNMTTPNKLLRLEDDGTLLYTMRLTISAECPMQLEDF
PMDAHACPLKFGSYAYPNSEVVYVWTNGSTKSVVVAEDGSRLNQYHLMGQTVGTENISTS
TGEYTIMTAHFHLKRKIGYFVIQTYLPCIMTVILSQVSFWLNRESVPARTVFGVTTVLTM
TTLSISARNSLPKVAYATAMDWFIAVCYAFVFSALIEFATVNYFTKRGWAWDGKKALEAA
KIKKKREVILNKSTNAFTTGKMSHPPNIPKEQTPAGTSNTTSVSVKPSEEKTSESKKTYN
SISKIDKMSRIVFPVLFGTFNLVYWATYLNREPVIKGAASPK
|
|
|
|---|
| BDBM50144862 |
|---|
| n/a |
|---|
| Name | BDBM50144862 |
|---|
| Synonyms: | 6-methyl-2-pyridyl 6-phenyl-4,5,7,8-tetraazatetracyclo[9.2.2.02,10.03,7]pentadeca-2(10),3,5,8-tetraen-9-yl ether | 6-methyl-2-pyridyl[6-phenyl-4,5,7,8-tetraazatetracyclo[9.2.2.02,10.03,7]pentadeca-2(10),3,5,8-tetraen-9-yloxy]methane | CHEMBL68712 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H23N5O |
|---|
| Mol. Mass. | 397.4723 |
|---|
| SMILES | Cc1cccc(COc2nn3c(nnc3c3C4CCC(CC4)c23)-c2ccccc2)n1 |(4.36,-17.37,;3.02,-16.61,;1.69,-17.39,;.36,-16.62,;.36,-15.08,;1.69,-14.31,;1.69,-12.77,;.36,-12,;.36,-10.46,;1.69,-9.69,;1.69,-8.15,;2.83,-7.12,;2.2,-5.7,;.68,-5.88,;.36,-7.38,;-.97,-8.17,;-2.3,-7.38,;-3.08,-8.71,;-1.54,-9.13,;-2.3,-10.46,;-3.63,-9.71,;-3.63,-8.17,;-.97,-9.71,;4.35,-7.43,;5.35,-6.28,;6.87,-6.6,;7.35,-8.06,;6.32,-9.22,;4.81,-8.9,;3.02,-15.08,)| |
|---|
| Structure | 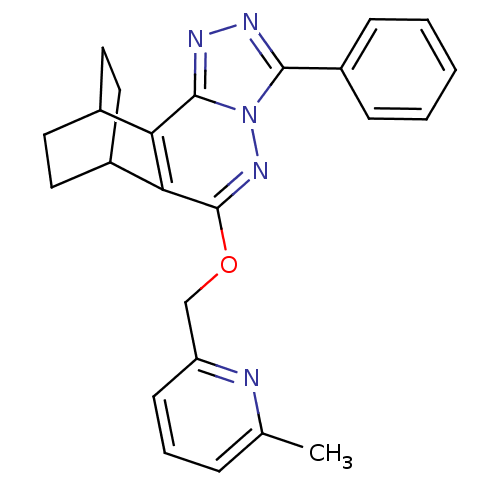
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Street, LJ; Sternfeld, F; Jelley, RA; Reeve, AJ; Carling, RW; Moore, KW; McKernan, RM; Sohal, B; Cook, S; Pike, A; Dawson, GR; Bromidge, FA; Wafford, KA; Seabrook, GR; Thompson, SA; Marshall, G; Pillai, GV; Castro, JL; Atack, JR; MacLeod, AM Synthesis and biological evaluation of 3-heterocyclyl-7,8,9,10-tetrahydro-(7,10-ethano)-1,2,4-triazolo[3,4-a]phthalazines and analogues as subtype-selective inverse agonists for the GABA(A)alpha5 benzodiazepine binding site. J Med Chem47:3642-57 (2004) [PubMed] Article
Street, LJ; Sternfeld, F; Jelley, RA; Reeve, AJ; Carling, RW; Moore, KW; McKernan, RM; Sohal, B; Cook, S; Pike, A; Dawson, GR; Bromidge, FA; Wafford, KA; Seabrook, GR; Thompson, SA; Marshall, G; Pillai, GV; Castro, JL; Atack, JR; MacLeod, AM Synthesis and biological evaluation of 3-heterocyclyl-7,8,9,10-tetrahydro-(7,10-ethano)-1,2,4-triazolo[3,4-a]phthalazines and analogues as subtype-selective inverse agonists for the GABA(A)alpha5 benzodiazepine binding site. J Med Chem47:3642-57 (2004) [PubMed] Article