

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cytochrome P450 3A4 | ||
| Ligand | BDBM50585945 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2164346 (CHEMBL5049207) | ||
| IC50 | 5700±n/a nM | ||
| Citation |  Poddutoori, R; Aardalen, K; Aithal, K; Barahagar, SS; Belliappa, C; Bock, M; Chelur, S; Gerken, A; Gopinath, S; Gruenenfelder, B; Kiffe, M; Krishnaswami, M; Langowski, J; Madapa, S; Narayanan, K; Pandit, C; Panigrahi, SK; Perrone, M; Potakamuri, RK; Ramachandra, M; Ramanathan, A; Ramos, R; Sager, E; Samajdar, S; Subramanya, HS; Thimmasandra, DS; Venetsanakos, E; M÷bitz, H Discovery of MAP855, an Efficacious and Selective MEK1/2 Inhibitor with an ATP-Competitive Mode of Action. J Med Chem65:4350-4366 (2022) [PubMed] Article Poddutoori, R; Aardalen, K; Aithal, K; Barahagar, SS; Belliappa, C; Bock, M; Chelur, S; Gerken, A; Gopinath, S; Gruenenfelder, B; Kiffe, M; Krishnaswami, M; Langowski, J; Madapa, S; Narayanan, K; Pandit, C; Panigrahi, SK; Perrone, M; Potakamuri, RK; Ramachandra, M; Ramanathan, A; Ramos, R; Sager, E; Samajdar, S; Subramanya, HS; Thimmasandra, DS; Venetsanakos, E; M÷bitz, H Discovery of MAP855, an Efficacious and Selective MEK1/2 Inhibitor with an ATP-Competitive Mode of Action. J Med Chem65:4350-4366 (2022) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cytochrome P450 3A4 | |||
| Name: | Cytochrome P450 3A4 | ||
| Synonyms: | Albendazole monooxygenase | Albendazole sulfoxidase | CP3A4_HUMAN | CYP3A3 | CYP3A4 | CYPIIIA3 | CYPIIIA4 | Cytochrome P450 3A3 | Cytochrome P450 3A4 (CYP3A4) | Cytochrome P450 HLp | Nifedipine oxidase | Quinine 3-monooxygenase | Taurochenodeoxycholate 6-alpha-hydroxylase | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 57349.57 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 503 | ||
| Sequence: |
| ||
| BDBM50585945 | |||
| n/a | |||
| Name | BDBM50585945 | ||
| Synonyms: | CHEMBL5088062 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C28H23ClFN7O | ||
| Mol. Mass. | 527.98 | ||
| SMILES | N[C@]1(CC#N)CC[C@@H](CC1)n1cnc2cnc3cc(F)c(cc3c12)-c1ccc(Oc2ncccn2)cc1Cl |r,wU:7.10,1.0,(-2.87,-4.67,;-2.48,-3.19,;-1.39,-4.27,;.1,-3.88,;1.59,-3.48,;-2.08,-1.7,;-3.17,-.61,;-4.65,-1.01,;-5.05,-2.5,;-3.96,-3.58,;-5.74,.08,;-7.28,-.08,;-7.9,1.33,;-6.76,2.36,;-6.76,3.89,;-5.43,4.67,;-4.09,3.9,;-2.77,4.66,;-1.43,3.9,;-.1,4.67,;-1.43,2.36,;-2.76,1.59,;-4.09,2.36,;-5.42,1.59,;-.1,1.59,;-.1,.04,;1.24,-.72,;2.57,.05,;3.9,-.72,;5.23,.05,;5.23,1.59,;6.56,2.35,;7.9,1.58,;7.9,.05,;6.57,-.73,;2.57,1.59,;1.24,2.36,;1.24,3.9,)| | ||
| Structure | 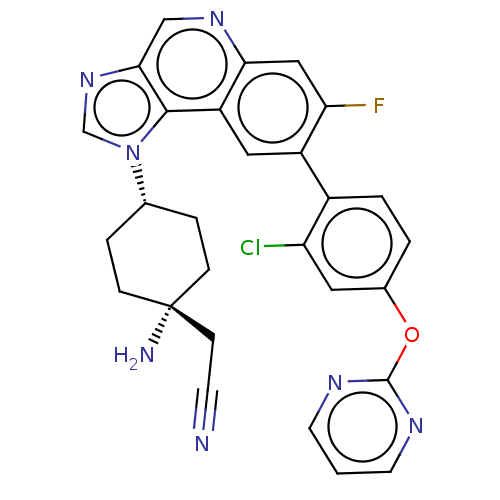
| ||