| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prolyl endopeptidase FAP |
|---|
| Ligand | BDBM50601879 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2238500 (CHEMBL5152396) |
|---|
| Ki | 720±n/a nM |
|---|
| Citation |  Plescia, J; Hédou, D; Pousse, ME; Labarre, A; Dufresne, C; Mittermaier, A; Moitessier, N Modulating the selectivity of inhibitors for prolyl oligopeptidase inhibitors and fibroblast activation protein-? for different indications. Eur J Med Chem240:0 (2022) [PubMed] Article Plescia, J; Hédou, D; Pousse, ME; Labarre, A; Dufresne, C; Mittermaier, A; Moitessier, N Modulating the selectivity of inhibitors for prolyl oligopeptidase inhibitors and fibroblast activation protein-? for different indications. Eur J Med Chem240:0 (2022) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Prolyl endopeptidase FAP |
|---|
| Name: | Prolyl endopeptidase FAP |
|---|
| Synonyms: | 170 kDa melanoma membrane-bound gelatinase | FAP | Fibroblast Activation Protein (FAP) | Fibroblast activation protein alpha | Integral membrane serine protease | SEPR_HUMAN | Seprase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 87712.48 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q12884 |
|---|
| Residue: | 760 |
|---|
| Sequence: | MKTWVKIVFGVATSAVLALLVMCIVLRPSRVHNSEENTMRALTLKDILNGTFSYKTFFPN
WISGQEYLHQSADNNIVLYNIETGQSYTILSNRTMKSVNASNYGLSPDRQFVYLESDYSK
LWRYSYTATYYIYDLSNGEFVRGNELPRPIQYLCWSPVGSKLAYVYQNNIYLKQRPGDPP
FQITFNGRENKIFNGIPDWVYEEEMLATKYALWWSPNGKFLAYAEFNDTDIPVIAYSYYG
DEQYPRTINIPYPKAGAKNPVVRIFIIDTTYPAYVGPQEVPVPAMIASSDYYFSWLTWVT
DERVCLQWLKRVQNVSVLSICDFREDWQTWDCPKTQEHIEESRTGWAGGFFVSTPVFSYD
AISYYKIFSDKDGYKHIHYIKDTVENAIQITSGKWEAINIFRVTQDSLFYSSNEFEEYPG
RRNIYRISIGSYPPSKKCVTCHLRKERCQYYTASFSDYAKYYALVCYGPGIPISTLHDGR
TDQEIKILEENKELENALKNIQLPKEEIKKLEVDEITLWYKMILPPQFDRSKKYPLLIQV
YGGPCSQSVRSVFAVNWISYLASKEGMVIALVDGRGTAFQGDKLLYAVYRKLGVYEVEDQ
ITAVRKFIEMGFIDEKRIAIWGWSYGGYVSSLALASGTGLFKCGIAVAPVSSWEYYASVY
TERFMGLPTKDDNLEHYKNSTVMARAEYFRNVDYLLIHGTADDNVHFQNSAQIAKALVNA
QVDFQAMWYSDQNHGLSGLSTNHLYTHMTHFLKQCFSLSD
|
|
|
|---|
| BDBM50601879 |
|---|
| n/a |
|---|
| Name | BDBM50601879 |
|---|
| Synonyms: | CHEMBL5199424 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C8H15BN2O4 |
|---|
| Mol. Mass. | 214.027 |
|---|
| SMILES | [H][C@]12CC[C@@H](B(O)O)N1C(=O)[C@@H](N)[C@@H](C)O2 |r| |
|---|
| Structure | 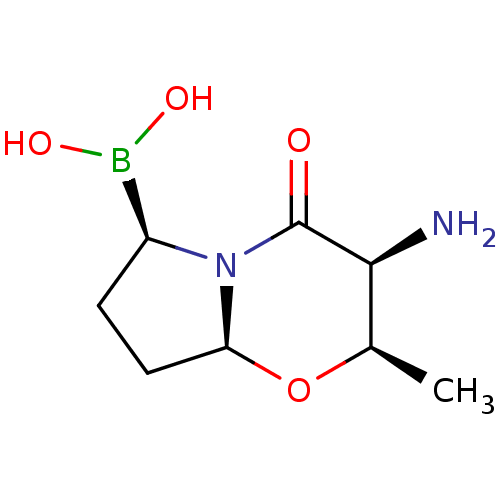
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Plescia, J; Hédou, D; Pousse, ME; Labarre, A; Dufresne, C; Mittermaier, A; Moitessier, N Modulating the selectivity of inhibitors for prolyl oligopeptidase inhibitors and fibroblast activation protein-? for different indications. Eur J Med Chem240:0 (2022) [PubMed] Article
Plescia, J; Hédou, D; Pousse, ME; Labarre, A; Dufresne, C; Mittermaier, A; Moitessier, N Modulating the selectivity of inhibitors for prolyl oligopeptidase inhibitors and fibroblast activation protein-? for different indications. Eur J Med Chem240:0 (2022) [PubMed] Article