| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 3-dehydroquinate dehydratase |
|---|
| Ligand | BDBM14672 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_332876 (CHEMBL858936) |
|---|
| Kd | 2500±n/a nM |
|---|
| Citation |  Robinson, DA; Stewart, KA; Price, NC; Chalk, PA; Coggins, JR; Lapthorn, AJ Crystal structures of Helicobacter pylori type II dehydroquinase inhibitor complexes: new directions for inhibitor design. J Med Chem49:1282-90 (2006) [PubMed] Article Robinson, DA; Stewart, KA; Price, NC; Chalk, PA; Coggins, JR; Lapthorn, AJ Crystal structures of Helicobacter pylori type II dehydroquinase inhibitor complexes: new directions for inhibitor design. J Med Chem49:1282-90 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 3-dehydroquinate dehydratase |
|---|
| Name: | 3-dehydroquinate dehydratase |
|---|
| Synonyms: | AROQ_HELPY | aroQ |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 18479.08 |
|---|
| Organism: | Helicobacter pylori |
|---|
| Description: | ChEMBL_766424 |
|---|
| Residue: | 167 |
|---|
| Sequence: | MKILVIQGPNLNMLGHRDPRLYGMVTLDQIHEIMQTFVKQGNLDVELEFFQTNFEGEIID
KIQESVGSDYEGIIINPGAFSHTSIAIADAIMLAGKPVIEVHLTNIQAREEFRKNSYTGA
ACGGVIMGFGPLGYNMALMAMVNILAEMKAFQEAQKNNPNNPINNQK
|
|
|
|---|
| BDBM14672 |
|---|
| n/a |
|---|
| Name | BDBM14672 |
|---|
| Synonyms: | 2-hydroxypropane-1,2,3-tricarboxylic acid | CHEMBL1261 | CITRIC ACID | Citrate | Citric Acid | Fragment 2 | cid_311 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C6H8O7 |
|---|
| Mol. Mass. | 192.1235 |
|---|
| SMILES | OC(=O)CC(O)(CC(O)=O)C(O)=O |
|---|
| Structure | 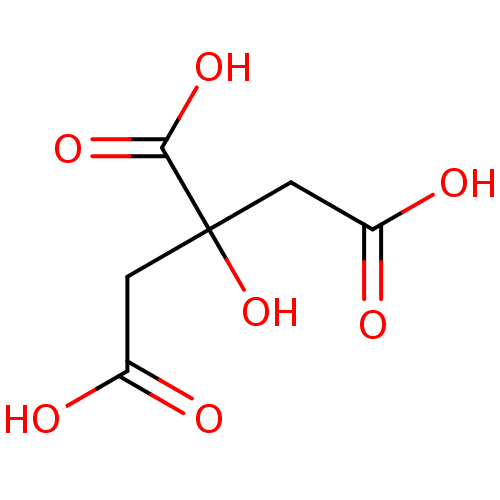
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Robinson, DA; Stewart, KA; Price, NC; Chalk, PA; Coggins, JR; Lapthorn, AJ Crystal structures of Helicobacter pylori type II dehydroquinase inhibitor complexes: new directions for inhibitor design. J Med Chem49:1282-90 (2006) [PubMed] Article
Robinson, DA; Stewart, KA; Price, NC; Chalk, PA; Coggins, JR; Lapthorn, AJ Crystal structures of Helicobacter pylori type II dehydroquinase inhibitor complexes: new directions for inhibitor design. J Med Chem49:1282-90 (2006) [PubMed] Article