| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Receptor-interacting serine/threonine-protein kinase 1 |
|---|
| Ligand | BDBM50322535 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2239009 (CHEMBL5152905) |
|---|
| IC50 | 12±n/a nM |
|---|
| Citation |  Shi, K; Zhang, J; Zhou, E; Wang, J; Wang, Y Small-Molecule Receptor-Interacting Protein 1 (RIP1) Inhibitors as Therapeutic Agents for Multifaceted Diseases: Current Medicinal Chemistry Insights and Emerging Opportunities. J Med Chem65:14971-14999 (2022) [PubMed] Article Shi, K; Zhang, J; Zhou, E; Wang, J; Wang, Y Small-Molecule Receptor-Interacting Protein 1 (RIP1) Inhibitors as Therapeutic Agents for Multifaceted Diseases: Current Medicinal Chemistry Insights and Emerging Opportunities. J Med Chem65:14971-14999 (2022) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Receptor-interacting serine/threonine-protein kinase 1 |
|---|
| Name: | Receptor-interacting serine/threonine-protein kinase 1 |
|---|
| Synonyms: | Cell death protein RIP | RIP | RIP-1 | RIP1 | RIPK1 | RIPK1_HUMAN | Receptor-interacting protein 1 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 75926.99 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q13546 |
|---|
| Residue: | 671 |
|---|
| Sequence: | MQPDMSLNVIKMKSSDFLESAELDSGGFGKVSLCFHRTQGLMIMKTVYKGPNCIEHNEAL
LEEAKMMNRLRHSRVVKLLGVIIEEGKYSLVMEYMEKGNLMHVLKAEMSTPLSVKGRIIL
EIIEGMCYLHGKGVIHKDLKPENILVDNDFHIKIADLGLASFKMWSKLNNEEHNELREVD
GTAKKNGGTLYYMAPEHLNDVNAKPTEKSDVYSFAVVLWAIFANKEPYENAICEQQLIMC
IKSGNRPDVDDITEYCPREIISLMKLCWEANPEARPTFPGIEEKFRPFYLSQLEESVEED
VKSLKKEYSNENAVVKRMQSLQLDCVAVPSSRSNSATEQPGSLHSSQGLGMGPVEESWFA
PSLEHPQEENEPSLQSKLQDEANYHLYGSRMDRQTKQQPRQNVAYNREEERRRRVSHDPF
AQQRPYENFQNTEGKGTAYSSAASHGNAVHQPSGLTSQPQVLYQNNGLYSSHGFGTRPLD
PGTAGPRVWYRPIPSHMPSLHNIPVPETNYLGNTPTMPFSSLPPTDESIKYTIYNSTGIQ
IGAYNYMEIGGTSSSLLDSTNTNFKEEPAAKYQAIFDNTTSLTDKHLDPIRENLGKHWKN
CARKLGFTQSQIDEIDHDYERDGLKEKVYQMLQKWVMREGIKGATVGKLAQALHQCSRID
LLSSLIYVSQN
|
|
|
|---|
| BDBM50322535 |
|---|
| n/a |
|---|
| Name | BDBM50322535 |
|---|
| Synonyms: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide | 3-[2-(Imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide | CHEMBL1171837 | PONATINIB | US10464902, Ponatinib | US9255107, AP24534 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C29H27F3N6O |
|---|
| Mol. Mass. | 532.5595 |
|---|
| SMILES | CN1CCN(Cc2ccc(NC(=O)c3ccc(C)c(c3)C#Cc3cnc4cccnn34)cc2C(F)(F)F)CC1 |
|---|
| Structure | 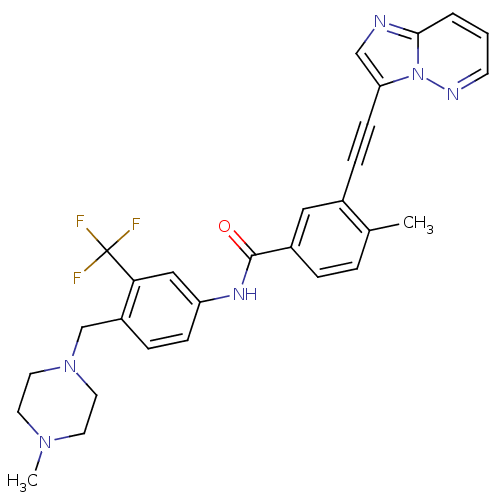
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Shi, K; Zhang, J; Zhou, E; Wang, J; Wang, Y Small-Molecule Receptor-Interacting Protein 1 (RIP1) Inhibitors as Therapeutic Agents for Multifaceted Diseases: Current Medicinal Chemistry Insights and Emerging Opportunities. J Med Chem65:14971-14999 (2022) [PubMed] Article
Shi, K; Zhang, J; Zhou, E; Wang, J; Wang, Y Small-Molecule Receptor-Interacting Protein 1 (RIP1) Inhibitors as Therapeutic Agents for Multifaceted Diseases: Current Medicinal Chemistry Insights and Emerging Opportunities. J Med Chem65:14971-14999 (2022) [PubMed] Article