| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Genome polyprotein |
|---|
| Ligand | BDBM50187935 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_388024 (CHEMBL869782) |
|---|
| Ki | 25±n/a nM |
|---|
| Citation |  Arasappan, A; Njoroge, FG; Chen, KX; Venkatraman, S; Parekh, TN; Gu, H; Pichardo, J; Butkiewicz, N; Prongay, A; Madison, V; Girijavallabhan, V P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease. Bioorg Med Chem Lett16:3960-5 (2006) [PubMed] Article Arasappan, A; Njoroge, FG; Chen, KX; Venkatraman, S; Parekh, TN; Gu, H; Pichardo, J; Butkiewicz, N; Prongay, A; Madison, V; Girijavallabhan, V P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease. Bioorg Med Chem Lett16:3960-5 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Genome polyprotein |
|---|
| Name: | Genome polyprotein |
|---|
| Synonyms: | Hepatitis C virus NS3 protease/helicase | Hepatitis C virus serine protease, NS3/NS4A |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 67067.41 |
|---|
| Organism: | Hepatitis C virus |
|---|
| Description: | A3EZI9 |
|---|
| Residue: | 631 |
|---|
| Sequence: | APITAYAQQTRGLLGCIITSLTGRDKNQVEGEVQIVSTAAQTFLATCINGVCWTVYHGAG
TRTIASSKGPVIQMYTNVDQDLVGWPAPQGARSLTPCTCGSSDLYLVTRHADVIPVRRRG
DGRGSLLSPRPISYLKGSSGGPLLCPAGHAVGIFRAAVCTRGVAKAVDFIPVEGLETTMR
SPVFSDNSSPPAVPQSYQVAHLHAPTGSGKSTKVPAAYAAQGYKVLVLNPSVAATLGFGA
YMSKAHGIDPNIRTGVRTITTGSPITYSTYGKFLADGGCSGGAYDIIICDECHSTDATSI
LGIGTVLDQAETAGARLTVLATATPPGSVTVPHPNIEEVALSTTGEIPFYGKAIPLEAIK
GGRHLIFCHSKKKCDELAAKLVALGVNAVAYYRGLDVSVIPASGDVVVVATDALMTGFTG
DFDSVIDCNTCVTQTVDFSLDPTFTIETTTLPQDAVSRTQRRGRTGRGKPGIYRFVTPGE
RPSGMFDSSVLCECYDAGCAWYELTPAETTVRLRAYMNTPGLPVCQDHLEFWEGVFTGLT
HIDAHFLSQTKQSGENLPYLVAYQATVCARAQAPPPSWDQMWKCLIRLKPTLHGPTPLLY
RLGAVQNEITLTHPITKYIMTCMSADLEVVT
|
|
|
|---|
| BDBM50187935 |
|---|
| n/a |
|---|
| Name | BDBM50187935 |
|---|
| Synonyms: | (6R,8S,11S)-11-cyclohexyl-10,13-dioxo-2,5-dioxa-9,12-diaza-tricyclo[14.3.1.1*6,9*]henicosa-1(19),16(20),17-triene-8-carboxylic acid [1-((S)-dimethylcarbamoyl-phenyl-methyl)-carbamoyl]-methyl)-aminooxalyl)-butyl)-amide | CHEMBL211701 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C42H56N6O9 |
|---|
| Mol. Mass. | 788.9288 |
|---|
| SMILES | CCCC(NC(=O)[C@@H]1C[C@@H]2CN1C(=O)[C@@H](NC(=O)CCc1cccc(OCCO2)c1)C1CCCCC1)C(=O)C(=O)NCC(=O)N[C@H](C(=O)N(C)C)c1ccccc1 |
|---|
| Structure | 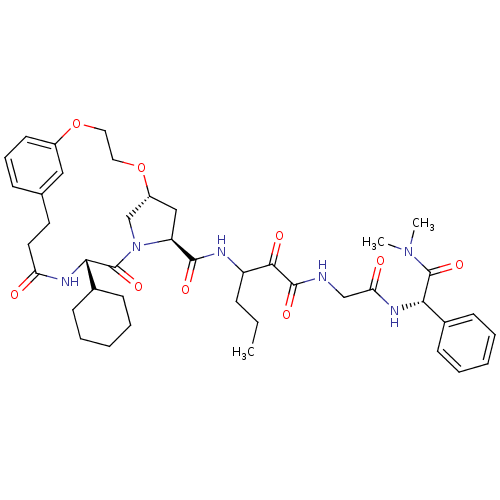
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Arasappan, A; Njoroge, FG; Chen, KX; Venkatraman, S; Parekh, TN; Gu, H; Pichardo, J; Butkiewicz, N; Prongay, A; Madison, V; Girijavallabhan, V P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease. Bioorg Med Chem Lett16:3960-5 (2006) [PubMed] Article
Arasappan, A; Njoroge, FG; Chen, KX; Venkatraman, S; Parekh, TN; Gu, H; Pichardo, J; Butkiewicz, N; Prongay, A; Madison, V; Girijavallabhan, V P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease. Bioorg Med Chem Lett16:3960-5 (2006) [PubMed] Article