| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor delta |
|---|
| Ligand | BDBM50188025 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_389973 (CHEMBL868636) |
|---|
| EC50 | 2400±n/a nM |
|---|
| Citation |  Kuhn, B; Hilpert, H; Benz, J; Binggeli, A; Grether, U; Humm, R; Märki, HP; Meyer, M; Mohr, P Structure-based design of indole propionic acids as novel PPARalpha/gamma co-agonists. Bioorg Med Chem Lett16:4016-20 (2006) [PubMed] Article Kuhn, B; Hilpert, H; Benz, J; Binggeli, A; Grether, U; Humm, R; Märki, HP; Meyer, M; Mohr, P Structure-based design of indole propionic acids as novel PPARalpha/gamma co-agonists. Bioorg Med Chem Lett16:4016-20 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor delta |
|---|
| Name: | Peroxisome proliferator-activated receptor delta |
|---|
| Synonyms: | NR1C2 | NUC1 | NUCI | Nuclear hormone receptor 1 | Nuclear receptor subfamily 1 group C member 2 | PPAR delta | PPAR-beta | PPARB | PPARD | PPARD_HUMAN | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor beta | Peroxisome proliferator-activated receptor delta |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 49910.45 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q03181 |
|---|
| Residue: | 441 |
|---|
| Sequence: | MEQPQEEAPEVREEEEKEEVAEAEGAPELNGGPQHALPSSSYTDLSRSSSPPSLLDQLQM
GCDGASCGSLNMECRVCGDKASGFHYGVHACEGCKGFFRRTIRMKLEYEKCERSCKIQKK
NRNKCQYCRFQKCLALGMSHNAIRFGRMPEAEKRKLVAGLTANEGSQYNPQVADLKAFSK
HIYNAYLKNFNMTKKKARSILTGKASHTAPFVIHDIETLWQAEKGLVWKQLVNGLPPYKE
ISVHVFYRCQCTTVETVRELTEFAKSIPSFSSLFLNDQVTLLKYGVHEAIFAMLASIVNK
DGLLVANGSGFVTREFLRSLRKPFSDIIEPKFEFAVKFNALELDDSDLALFIAAIILCGD
RPGLMNVPRVEAIQDTILRALEFHLQANHPDAQYLFPKLLQKMADLRQLVTEHAQMMQRI
KKTETETSLHPLLQEIYKDMY
|
|
|
|---|
| BDBM50188025 |
|---|
| n/a |
|---|
| Name | BDBM50188025 |
|---|
| Synonyms: | 3-(1-((2-(3,5-dimethoxyphenyl)-5-methyloxazol-4-yl)methyl)-1H-indol-5-yl)-2-ethoxypropanoic acid | CHEMBL211839 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H28N2O6 |
|---|
| Mol. Mass. | 464.5103 |
|---|
| SMILES | CCOC(Cc1ccc2n(Cc3nc(oc3C)-c3cc(OC)cc(OC)c3)ccc2c1)C(O)=O |
|---|
| Structure | 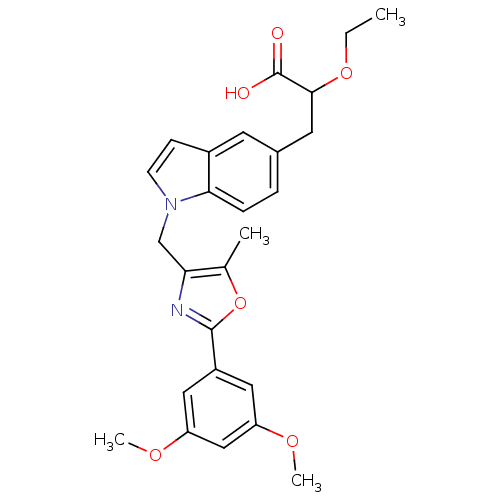
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Kuhn, B; Hilpert, H; Benz, J; Binggeli, A; Grether, U; Humm, R; Märki, HP; Meyer, M; Mohr, P Structure-based design of indole propionic acids as novel PPARalpha/gamma co-agonists. Bioorg Med Chem Lett16:4016-20 (2006) [PubMed] Article
Kuhn, B; Hilpert, H; Benz, J; Binggeli, A; Grether, U; Humm, R; Märki, HP; Meyer, M; Mohr, P Structure-based design of indole propionic acids as novel PPARalpha/gamma co-agonists. Bioorg Med Chem Lett16:4016-20 (2006) [PubMed] Article