| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Equilibrative nucleoside transporter 2 |
|---|
| Ligand | BDBM56941 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2261598 (CHEMBL5216609) |
|---|
| IC50 | 570±n/a nM |
|---|
| Citation |  Shahari, MSB; Dolzhenko, AV A closer look at N Eur J Med Chem241:0 (2022) [PubMed] Article Shahari, MSB; Dolzhenko, AV A closer look at N Eur J Med Chem241:0 (2022) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Equilibrative nucleoside transporter 2 |
|---|
| Name: | Equilibrative nucleoside transporter 2 |
|---|
| Synonyms: | 36 kDa nucleolar protein HNP36 | DER12 | DER12 | Delayed-early response protein 12 | ENT2 | Equilibrative NBMPR-insensitive nucleoside transporter | Equilibrative nitrobenzylmercaptopurine riboside-insensitive nucleoside transporter | Equilibrative nucleoside transporter 2 | HNP36 | Hydrophobic nucleolar protein, 36 kDa | Nucleoside transporter, ei-type | S29A2_HUMAN | SLC29A2 | Solute carrier family 29 member 2 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 50110.91 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_109695 |
|---|
| Residue: | 456 |
|---|
| Sequence: | MARGDAPRDSYHLVGISFFILGLGTLLPWNFFITAIPYFQARLAGAGNSTARILSTNHTG
PEDAFNFNNWVTLLSQLPLLLFTLLNSFLYQCVPETVRILGSLLAILLLFALTAALVKVD
MSPGPFFSITMASVCFINSFSAVLQGSLFGQLGTMPSTYSTLFLSGQGLAGIFAALAMLL
SMASGVDAETSALGYFITPCVGILMSIVCYLSLPHLKFARYYLANKSSQAQAQELETKAE
LLQSDENGIPSSPQKVALTLDLDLEKEPESEPDEPQKPGKPSVFTVFQKIWLTALCLVLV
FTVTLSVFPAITAMVTSSTSPGKWSQFFNPICCFLLFNIMDWLGRSLTSYFLWPDEDSRL
LPLLVCLRFLFVPLFMLCHVPQRSRLPILFPQDAYFITFMLLFAVSNGYLVSLTMCLAPR
QVLPHEREVAGALMTFFLALGLSCGASLSFLFKALL
|
|
|
|---|
| BDBM56941 |
|---|
| n/a |
|---|
| Name | BDBM56941 |
|---|
| Synonyms: | 6-[[4-(4-fluorophenyl)-1-piperazinyl]methyl]-N2-(3-methylphenyl)-1,3,5-triazine-2,4-diamine | 6-[[4-(4-fluorophenyl)piperazin-1-yl]methyl]-2-N-(3-methylphenyl)-1,3,5-triazine-2,4-diamine | 6-[[4-(4-fluorophenyl)piperazin-1-yl]methyl]-N2-(3-methylphenyl)-1,3,5-triazine-2,4-diamine | MLS000084209 | SMR000047961 | [4-amino-6-[[4-(4-fluorophenyl)piperazino]methyl]-s-triazin-2-yl]-(m-tolyl)amine | cid_666635 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H24FN7 |
|---|
| Mol. Mass. | 393.4606 |
|---|
| SMILES | Cc1cccc(Nc2nc(N)nc(CN3CCN(CC3)c3ccc(F)cc3)n2)c1 |
|---|
| Structure | 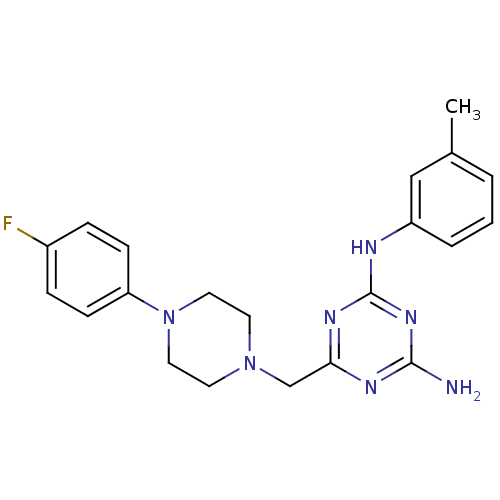
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Shahari, MSB; Dolzhenko, AV A closer look at N Eur J Med Chem241:0 (2022) [PubMed] Article
Shahari, MSB; Dolzhenko, AV A closer look at N Eur J Med Chem241:0 (2022) [PubMed] Article