

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | 11-beta-hydroxysteroid dehydrogenase type 2 | ||
| Ligand | BDBM50202092 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_423359 (CHEMBL909906) | ||
| IC50 | >20000±n/a nM | ||
| Citation |  Rohde, JJ; Pliushchev, MA; Sorensen, BK; Wodka, D; Shuai, Q; Wang, J; Fung, S; Monzon, KM; Chiou, WJ; Pan, L; Deng, X; Chovan, LE; Ramaiya, A; Mullally, M; Henry, RF; Stolarik, DF; Imade, HM; Marsh, KC; Beno, DW; Fey, TA; Droz, BA; Brune, ME; Camp, HS; Sham, HL; Frevert, EU; Jacobson, PB; Link, JT Discovery and metabolic stabilization of potent and selective 2-amino-N-(adamant-2-yl) acetamide 11beta-hydroxysteroid dehydrogenase type 1 inhibitors. J Med Chem50:149-64 (2007) [PubMed] Article Rohde, JJ; Pliushchev, MA; Sorensen, BK; Wodka, D; Shuai, Q; Wang, J; Fung, S; Monzon, KM; Chiou, WJ; Pan, L; Deng, X; Chovan, LE; Ramaiya, A; Mullally, M; Henry, RF; Stolarik, DF; Imade, HM; Marsh, KC; Beno, DW; Fey, TA; Droz, BA; Brune, ME; Camp, HS; Sham, HL; Frevert, EU; Jacobson, PB; Link, JT Discovery and metabolic stabilization of potent and selective 2-amino-N-(adamant-2-yl) acetamide 11beta-hydroxysteroid dehydrogenase type 1 inhibitors. J Med Chem50:149-64 (2007) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| 11-beta-hydroxysteroid dehydrogenase type 2 | |||
| Name: | 11-beta-hydroxysteroid dehydrogenase type 2 | ||
| Synonyms: | 11-DH2 | 11-beta-HSD2 | 11-beta-Hydroxysteroid Dehydrogenase 2 (11-beta-HSD2) | 11-beta-hydroxysteroid dehydrogenase | 11-beta-hydroxysteroid dehydrogenase 2 | 11-beta-hydroxysteroid dehydrogenase type 2 | 11-beta-hydroxysteroid dehydrogenase type 2 (11-beta-HSD2) | Corticosteroid 11-beta-dehydrogenase isozyme 2 | DHI2_HUMAN | HSD11B2 | HSD11K | NAD-dependent 11-beta-hydroxysteroid dehydrogenase | SDR9C3 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 44141.72 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Purified recombinant human 11beta-HSD2. | ||
| Residue: | 405 | ||
| Sequence: |
| ||
| BDBM50202092 | |||
| n/a | |||
| Name | BDBM50202092 | ||
| Synonyms: | CHEMBL386029 | N-[(E)-5-fluoro-2-adamantyl]-2-{4-[5-(trifluoromethyl)pyridin-2-yl]piperazin-1-yl}acetamide | ||
| Type | Small organic molecule | ||
| Emp. Form. | C22H28F4N4O | ||
| Mol. Mass. | 440.4775 | ||
| SMILES | FC(F)(F)c1ccc(nc1)N1CCN(CC(=O)N[C@H]2C3CC4CC2C[C@](F)(C4)C3)CC1 |r,wU:18.18,wD:25.27,TLB:18:19:27:22.23.24,17:18:27.21.22:24,THB:20:21:24:28.19.18,20:19:27.21.22:24,18:23:27:28.20.19,(7.79,-3.68,;6.48,-2.88,;5.67,-4.19,;7.31,-1.58,;5.18,-2.06,;3.82,-2.77,;2.52,-1.94,;2.59,-.41,;3.95,.31,;5.25,-.51,;1.29,.42,;-.05,-.34,;-1.37,.43,;-1.36,1.98,;-2.7,2.72,;-4.02,1.93,;-4,.39,;-5.37,2.68,;-6.69,1.89,;-6.77,.36,;-7.84,-.87,;-9.22,-.24,;-9.15,1.34,;-8.06,2.53,;-9.43,2.11,;-9.48,.63,;-10.85,1.23,;-10.74,-.6,;-8.18,.08,;-.04,2.74,;1.3,1.96,)| | ||
| Structure | 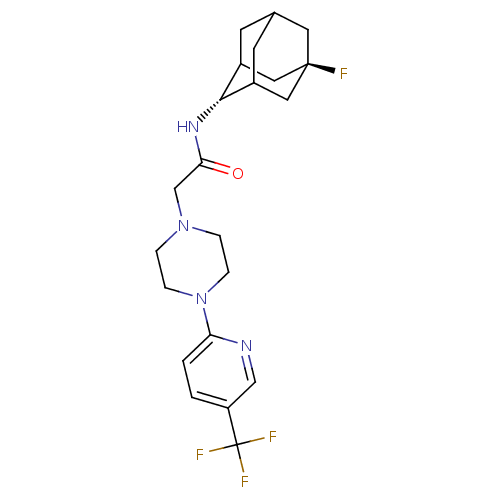
| ||