

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Prostaglandin G/H synthase 2 | ||
| Ligand | BDBM50207448 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_461151 (CHEMBL944174) | ||
| IC50 | 100±n/a nM | ||
| Citation |  Blobaum, AL; Marnett, LJ Structural and functional basis of cyclooxygenase inhibition. J Med Chem50:1425-41 (2007) [PubMed] Article Blobaum, AL; Marnett, LJ Structural and functional basis of cyclooxygenase inhibition. J Med Chem50:1425-41 (2007) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Prostaglandin G/H synthase 2 | |||
| Name: | Prostaglandin G/H synthase 2 | ||
| Synonyms: | COX2 | Cyclooxygenase | Cyclooxygenase 2 (COX-2) | Cyclooxygenase-2 | Cyclooxygenase-2 (COX-2 AA) | Cyclooxygenase-2 (COX-2 AEA) | Cyclooxygenase-2 (COX-2) | PGH synthase 2 | PGH2_HUMAN | PGHS-2 | PHS II | PTGS2 | Prostaglandin E synthase/G/H synthase 2 | Prostaglandin H2 synthase 2 | Prostaglandin-endoperoxide synthase 2 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 69003.89 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Recombinant Cox-2 provided by Cayman (Cayman Chemical Co.,Ann Arbor, MI). | ||
| Residue: | 604 | ||
| Sequence: |
| ||
| BDBM50207448 | |||
| n/a | |||
| Name | BDBM50207448 | ||
| Synonyms: | 2-(2',6'-diethoxy-2-fluoro-biphenyl4-yl)-propionic acid | CHEMBL257539 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C19H21FO4 | ||
| Mol. Mass. | 332.366 | ||
| SMILES | CCOc1cccc(OCC)c1-c1ccc(cc1F)C(C)C(O)=O |w:19.21,(26.76,-25.54,;28.1,-24.77,;29.43,-25.54,;30.76,-24.77,;30.77,-23.22,;32.1,-22.45,;33.43,-23.22,;33.44,-24.77,;34.77,-25.54,;36.1,-24.76,;37.44,-25.53,;32.1,-25.54,;32.1,-27.08,;30.76,-27.84,;30.76,-29.38,;32.1,-30.15,;33.44,-29.38,;33.43,-27.84,;34.76,-27.07,;32.1,-31.69,;30.77,-32.47,;33.43,-32.46,;33.44,-34.01,;34.77,-31.69,)| | ||
| Structure | 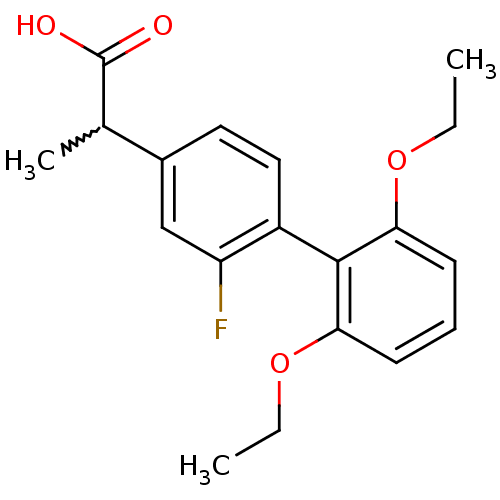
| ||