| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cytochrome P450 3A4 |
|---|
| Ligand | BDBM50208339 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_455108 (CHEMBL887138) |
|---|
| IC50 | 7000±n/a nM |
|---|
| Citation |  Feng, Y; Cameron, MD; Frackowiak, B; Griffin, E; Lin, L; Ruiz, C; Schröter, T; LoGrasso, P Structure-activity relationships, and drug metabolism and pharmacokinetic properties for indazole piperazine and indazole piperidine inhibitors of ROCK-II. Bioorg Med Chem Lett17:2355-60 (2007) [PubMed] Article Feng, Y; Cameron, MD; Frackowiak, B; Griffin, E; Lin, L; Ruiz, C; Schröter, T; LoGrasso, P Structure-activity relationships, and drug metabolism and pharmacokinetic properties for indazole piperazine and indazole piperidine inhibitors of ROCK-II. Bioorg Med Chem Lett17:2355-60 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cytochrome P450 3A4 |
|---|
| Name: | Cytochrome P450 3A4 |
|---|
| Synonyms: | Albendazole monooxygenase | Albendazole sulfoxidase | CP3A4_HUMAN | CYP3A3 | CYP3A4 | CYPIIIA3 | CYPIIIA4 | Cytochrome P450 3A3 | Cytochrome P450 3A4 (CYP3A4) | Cytochrome P450 HLp | Nifedipine oxidase | Quinine 3-monooxygenase | Taurochenodeoxycholate 6-alpha-hydroxylase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 57349.57 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 503 |
|---|
| Sequence: | MALIPDLAMETWLLLAVSLVLLYLYGTHSHGLFKKLGIPGPTPLPFLGNILSYHKGFCMF
DMECHKKYGKVWGFYDGQQPVLAITDPDMIKTVLVKECYSVFTNRRPFGPVGFMKSAISI
AEDEEWKRLRSLLSPTFTSGKLKEMVPIIAQYGDVLVRNLRREAETGKPVTLKDVFGAYS
MDVITSTSFGVNIDSLNNPQDPFVENTKKLLRFDFLDPFFLSITVFPFLIPILEVLNICV
FPREVTNFLRKSVKRMKESRLEDTQKHRVDFLQLMIDSQNSKETESHKALSDLELVAQSI
IFIFAGYETTSSVLSFIMYELATHPDVQQKLQEEIDAVLPNKAPPTYDTVLQMEYLDMVV
NETLRLFPIAMRLERVCKKDVEINGMFIPKGVVVMIPSYALHRDPKYWTEPEKFLPERFS
KKNKDNIDPYIYTPFGSGPRNCIGMRFALMNMKLALIRVLQNFSFKPCKETQIPLKLSLG
GLLQPEKPVVLKVESRDGTVSGA
|
|
|
|---|
| BDBM50208339 |
|---|
| n/a |
|---|
| Name | BDBM50208339 |
|---|
| Synonyms: | (R)-1-(4-(1H-indazol-5-yl)piperazin-1-yl)-2-amino-3-phenylpropan-1-one | CHEMBL240646 | SR-1357 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H23N5O |
|---|
| Mol. Mass. | 349.4295 |
|---|
| SMILES | N[C@H](Cc1ccccc1)C(=O)N1CCN(CC1)c1ccc2[nH]ncc2c1 |
|---|
| Structure | 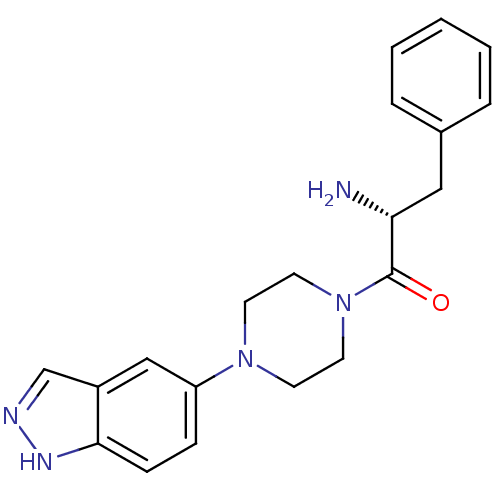
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Feng, Y; Cameron, MD; Frackowiak, B; Griffin, E; Lin, L; Ruiz, C; Schröter, T; LoGrasso, P Structure-activity relationships, and drug metabolism and pharmacokinetic properties for indazole piperazine and indazole piperidine inhibitors of ROCK-II. Bioorg Med Chem Lett17:2355-60 (2007) [PubMed] Article
Feng, Y; Cameron, MD; Frackowiak, B; Griffin, E; Lin, L; Ruiz, C; Schröter, T; LoGrasso, P Structure-activity relationships, and drug metabolism and pharmacokinetic properties for indazole piperazine and indazole piperidine inhibitors of ROCK-II. Bioorg Med Chem Lett17:2355-60 (2007) [PubMed] Article