| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Monoglyceride lipase |
|---|
| Ligand | BDBM50220343 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_446075 (CHEMBL895170) |
|---|
| IC50 | 73000±n/a nM |
|---|
| Citation |  Cisneros, JA; Vandevoorde, S; Ortega-Gutiérrez, S; Paris, C; Fowler, CJ; López-Rodríguez, ML Structure-activity relationship of a series of inhibitors of monoacylglycerol hydrolysis--comparison with effects upon fatty acid amide hydrolase. J Med Chem50:5012-23 (2007) [PubMed] Article Cisneros, JA; Vandevoorde, S; Ortega-Gutiérrez, S; Paris, C; Fowler, CJ; López-Rodríguez, ML Structure-activity relationship of a series of inhibitors of monoacylglycerol hydrolysis--comparison with effects upon fatty acid amide hydrolase. J Med Chem50:5012-23 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Monoglyceride lipase |
|---|
| Name: | Monoglyceride lipase |
|---|
| Synonyms: | MAGL | MGLL_RAT | Mgl2 | Mgll | Monoacylglycerol lipase | Monoglyceride lipase | Monoglyceride lipase (MGL) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 33505.38 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | Q8R431 |
|---|
| Residue: | 303 |
|---|
| Sequence: | MPEASSPRRTPQNVPYQDLPHLVNADGQYLFCRYWKPSGTPKALIFVSHGAGEHCGRYDE
LAQMLKRLDMLVFAHDHVGHGQSEGERMVVSDFQVFVRDLLQHVNTVQKDYPEVPVFLLG
HSMGGAISILAAAERPTHFSGMILISPLILANPESASTLKVLAAKLLNFVLPNISLGRID
SSVLSRNKSEVDLYNSDPLICHAGVKVCFGIQLLNAVSRVERAMPRLTLPFLLLQGSADR
LCDSKGAYLLMESSPSQDKTLKMYEGAYHVLHKELPEVTNSVLHEINTWVSHRIAVAGAR
CLP
|
|
|
|---|
| BDBM50220343 |
|---|
| n/a |
|---|
| Name | BDBM50220343 |
|---|
| Synonyms: | (+/-)-tetrahydro-2H-pyran-2-ylmethyl (9Z,12Z)-octadeca-9,12-dienoate | CHEMBL397182 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H42O3 |
|---|
| Mol. Mass. | 378.5885 |
|---|
| SMILES | CCCCC\C=C/C\C=C/CCCCCCCC(=O)OCC1CCCCO1 |w:21.20| |
|---|
| Structure | 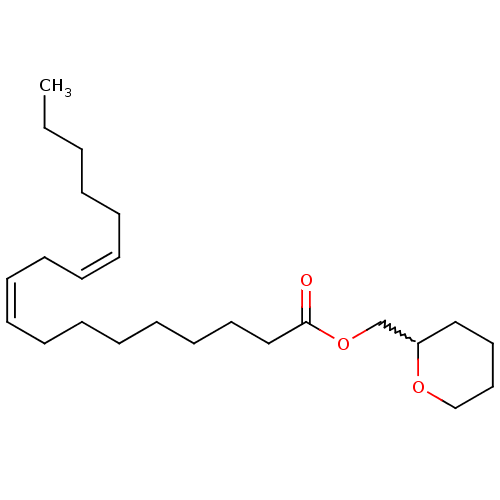
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Cisneros, JA; Vandevoorde, S; Ortega-Gutiérrez, S; Paris, C; Fowler, CJ; López-Rodríguez, ML Structure-activity relationship of a series of inhibitors of monoacylglycerol hydrolysis--comparison with effects upon fatty acid amide hydrolase. J Med Chem50:5012-23 (2007) [PubMed] Article
Cisneros, JA; Vandevoorde, S; Ortega-Gutiérrez, S; Paris, C; Fowler, CJ; López-Rodríguez, ML Structure-activity relationship of a series of inhibitors of monoacylglycerol hydrolysis--comparison with effects upon fatty acid amide hydrolase. J Med Chem50:5012-23 (2007) [PubMed] Article