| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 72 kDa type IV collagenase |
|---|
| Ligand | BDBM50229635 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_461864 (CHEMBL927857) |
|---|
| IC50 | 574±n/a nM |
|---|
| Citation |  Burns, DM; He, C; Li, Y; Scherle, P; Liu, X; Marando, CA; Covington, MB; Yang, G; Pan, M; Turner, S; Fridman, JS; Hollis, G; Vaddi, K; Yeleswaram, S; Newton, R; Friedman, S; Metcalf, B; Yao, W Conversion of an MMP-potent scaffold to an MMP-selective HER-2 sheddase inhibitor via scaffold hybridization and subtle P1' permutations. Bioorg Med Chem Lett18:560-4 (2008) [PubMed] Article Burns, DM; He, C; Li, Y; Scherle, P; Liu, X; Marando, CA; Covington, MB; Yang, G; Pan, M; Turner, S; Fridman, JS; Hollis, G; Vaddi, K; Yeleswaram, S; Newton, R; Friedman, S; Metcalf, B; Yao, W Conversion of an MMP-potent scaffold to an MMP-selective HER-2 sheddase inhibitor via scaffold hybridization and subtle P1' permutations. Bioorg Med Chem Lett18:560-4 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 72 kDa type IV collagenase |
|---|
| Name: | 72 kDa type IV collagenase |
|---|
| Synonyms: | 72 kDa gelatinase | 72 kDa type IV collagenase precursor | CLG4A | Gelatinase A | Gelatinase A (MMP-2) | MMP2 | MMP2_HUMAN | Matrix metalloproteinase-2 | Matrix metalloproteinase-2 (MMP 2) | Matrix metalloproteinase-2 (MMP2) | Matrix metalloproteinases 2 (MMP-2) | TBE-1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 73870.36 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P08253 |
|---|
| Residue: | 660 |
|---|
| Sequence: | MEALMARGALTGPLRALCLLGCLLSHAAAAPSPIIKFPGDVAPKTDKELAVQYLNTFYGC
PKESCNLFVLKDTLKKMQKFFGLPQTGDLDQNTIETMRKPRCGNPDVANYNFFPRKPKWD
KNQITYRIIGYTPDLDPETVDDAFARAFQVWSDVTPLRFSRIHDGEADIMINFGRWEHGD
GYPFDGKDGLLAHAFAPGTGVGGDSHFDDDELWTLGEGQVVRVKYGNADGEYCKFPFLFN
GKEYNSCTDTGRSDGFLWCSTTYNFEKDGKYGFCPHEALFTMGGNAEGQPCKFPFRFQGT
SYDSCTTEGRTDGYRWCGTTEDYDRDKKYGFCPETAMSTVGGNSEGAPCVFPFTFLGNKY
ESCTSAGRSDGKMWCATTANYDDDRKWGFCPDQGYSLFLVAAHEFGHAMGLEHSQDPGAL
MAPIYTYTKNFRLSQDDIKGIQELYGASPDIDLGTGPTPTLGPVTPEICKQDIVFDGIAQ
IRGEIFFFKDRFIWRTVTPRDKPMGPLLVATFWPELPEKIDAVYEAPQEEKAVFFAGNEY
WIYSASTLERGYPKPLTSLGLPPDVQRVDAAFNWSKNKKTYIFAGDKFWRYNEVKKKMDP
GFPKLIADAWNAIPDNLDAVVDLQGGGHSYFFKGAYYLKLENQSLKSVKFGSIKSDWLGC
|
|
|
|---|
| BDBM50229635 |
|---|
| n/a |
|---|
| Name | BDBM50229635 |
|---|
| Synonyms: | 4,4-dimethyl-tetrahydrofuran-3-yl 4-(hydroxycarbamoyl)-4-((4-o-tolylpiperidin-1-ylsulfonyl)methyl)piperidine-1-carboxylate | CHEMBL439405 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H39N3O7S |
|---|
| Mol. Mass. | 537.669 |
|---|
| SMILES | Cc1ccccc1C1CCN(CC1)S(=O)(=O)CC1(CCN(CC1)C(=O)OC1COCC1(C)C)C(=O)NO |w:26.28| |
|---|
| Structure | 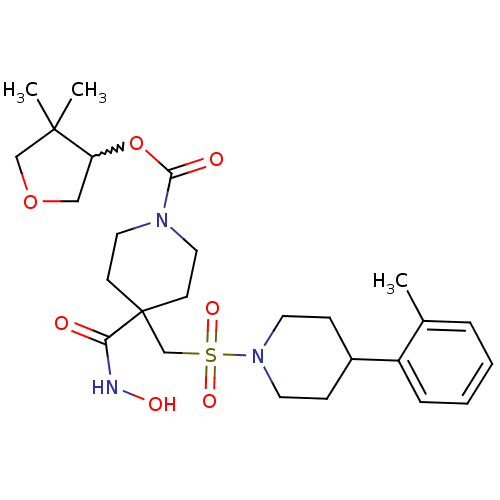
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Burns, DM; He, C; Li, Y; Scherle, P; Liu, X; Marando, CA; Covington, MB; Yang, G; Pan, M; Turner, S; Fridman, JS; Hollis, G; Vaddi, K; Yeleswaram, S; Newton, R; Friedman, S; Metcalf, B; Yao, W Conversion of an MMP-potent scaffold to an MMP-selective HER-2 sheddase inhibitor via scaffold hybridization and subtle P1' permutations. Bioorg Med Chem Lett18:560-4 (2008) [PubMed] Article
Burns, DM; He, C; Li, Y; Scherle, P; Liu, X; Marando, CA; Covington, MB; Yang, G; Pan, M; Turner, S; Fridman, JS; Hollis, G; Vaddi, K; Yeleswaram, S; Newton, R; Friedman, S; Metcalf, B; Yao, W Conversion of an MMP-potent scaffold to an MMP-selective HER-2 sheddase inhibitor via scaffold hybridization and subtle P1' permutations. Bioorg Med Chem Lett18:560-4 (2008) [PubMed] Article