| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Voltage-dependent T-type calcium channel subunit alpha-1I |
|---|
| Ligand | BDBM50240424 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_538845 (CHEMBL1033088) |
|---|
| IC50 | >5000000±n/a nM |
|---|
| Citation |  Yang, ZQ; Barrow, JC; Shipe, WD; Schlegel, KA; Shu, Y; Yang, FV; Lindsley, CW; Rittle, KE; Bock, MG; Hartman, GD; Uebele, VN; Nuss, CE; Fox, SV; Kraus, RL; Doran, SM; Connolly, TM; Tang, C; Ballard, JE; Kuo, Y; Adarayan, ED; Prueksaritanont, T; Zrada, MM; Marino, MJ; Graufelds, VK; DiLella, AG; Reynolds, IJ; Vargas, HM; Bunting, PB; Woltmann, RF; Magee, MM; Koblan, KS; Renger, JJ Discovery of 1,4-substituted piperidines as potent and selective inhibitors of T-type calcium channels. J Med Chem51:6471-7 (2008) [PubMed] Article Yang, ZQ; Barrow, JC; Shipe, WD; Schlegel, KA; Shu, Y; Yang, FV; Lindsley, CW; Rittle, KE; Bock, MG; Hartman, GD; Uebele, VN; Nuss, CE; Fox, SV; Kraus, RL; Doran, SM; Connolly, TM; Tang, C; Ballard, JE; Kuo, Y; Adarayan, ED; Prueksaritanont, T; Zrada, MM; Marino, MJ; Graufelds, VK; DiLella, AG; Reynolds, IJ; Vargas, HM; Bunting, PB; Woltmann, RF; Magee, MM; Koblan, KS; Renger, JJ Discovery of 1,4-substituted piperidines as potent and selective inhibitors of T-type calcium channels. J Med Chem51:6471-7 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Voltage-dependent T-type calcium channel subunit alpha-1I |
|---|
| Name: | Voltage-dependent T-type calcium channel subunit alpha-1I |
|---|
| Synonyms: | CAC1I_HUMAN | CACNA1I | KIAA1120 | Voltage-gated T-type calcium channel | Voltage-gated T-type calcium channel alpha-1I subunit |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 245101.25 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_538845 |
|---|
| Residue: | 2223 |
|---|
| Sequence: | MAESASPPSSSAAAPAAEPGVTTEQPGPRSPPSSPPGLEEPLDGADPHVPHPDLAPIAFF
CLRQTTSPRNWCIKMVCNPWFECVSMLVILLNCVTLGMYQPCDDMDCLSDRCKILQVFDD
FIFIFFAMEMVLKMVALGIFGKKCYLGDTWNRLDFFIVMAGMVEYSLDLQNINLSAIRTV
RVLRPLKAINRVPSMRILVNLLLDTLPMLGNVLLLCFFVFFIFGIIGVQLWAGLLRNRCF
LEENFTIQGDVALPPYYQPEEDDEMPFICSLSGDNGIMGCHEIPPLKEQGRECCLSKDDV
YDFGAGRQDLNASGLCVNWNRYYNVCRTGSANPHKGAINFDNIGYAWIVIFQVITLEGWV
EIMYYVMDAHSFYNFIYFILLIIVGSFFMINLCLVVIATQFSETKQREHRLMLEQRQRYL
SSSTVASYAEPGDCYEEIFQYVCHILRKAKRRALGLYQALQSRRQALGPEAPAPAKPGPH
AKEPRHYHGKTKGQGDEGRHLGSRHCQTLHGPASPGNDHSGRELCPQHSPLDATPHTLVQ
PIPATLASDPASCPCCQHEDGRRPSGLGSTDSGQEGSGSGSSAGGEDEADGDGARSSEDG
ASSELGKEEEEEEQADGAVWLCGDVWRETRAKLRGIVDSKYFNRGIMMAILVNTVSMGIE
HHEQPEELTNILEICNVVFTSMFALEMILKLAAFGLFDYLRNPYNIFDSIIVIISIWEIV
GQADGGLSVLRTFRLLRVLKLVRFMPALRRQLVVLMKTMDNVATFCMLLMLFIFIFSILG
MHIFGCKFSLRTDTGDTVPDRKNFDSLLWAIVTVFQILTQEDWNVVLYNGMASTSPWASL
YFVALMTFGNYVLFNLLVAILVEGFQAEGDANRSYSDEDQSSSNIEEFDKLQEGLDSSGD
PKLCPIPMTPNGHLDPSLPLGGHLGPAGAAGPAPRLSLQPDPMLVALGSRKSSVMSLGRM
SYDQRSLSSSRSSYYGPWGRSAAWASRRSSWNSLKHKPPSAEHESLLSAERGGGARVCEV
AADEGPPRAAPLHTPHAHHIHHGPHLAHRHRHHRRTLSLDNRDSVDLAELVPAVGAHPRA
AWRAAGPAPGHEDCNGRMPSIAKDVFTKMGDRGDRGEDEEEIDYTLCFRVRKMIDVYKPD
WCEVREDWSVYLFSPENRFRVLCQTIIAHKLFDYVVLAFIFLNCITIALERPQIEAGSTE
RIFLTVSNYIFTAIFVGEMTLKVVSLGLYFGEQAYLRSSWNVLDGFLVFVSIIDIVVSLA
SAGGAKILGVLRVLRLLRTLRPLRVISRAPGLKLVVETLISSLKPIGNIVLICCAFFIIF
GILGVQLFKGKFYHCLGVDTRNITNRSDCMAANYRWVHHKYNFDNLGQALMSLFVLASKD
GWVNIMYNGLDAVAVDQQPVTNHNPWMLLYFISFLLIVSFFVLNMFVGVVVENFHKCRQH
QEAEEARRREEKRLRRLEKKRRKAQRLPYYATYCHTRLLIHSMCTSHYLDIFITFIICLN
VVTMSLEHYNQPTSLETALKYCNYMFTTVFVLEAVLKLVAFGLRRFFKDRWNQLDLAIVL
LSVMGITLEEIEINAALPINPTIIRIMRVLRIARVLKLLKMATGMRALLDTVVQALPQVG
NLGLLFMLLFFIYAALGVELFGKLVCNDENPCEGMSRHATFENFGMAFLTLFQVSTGDNW
NGIMKDTLRDCTHDERSCLSSLQFVSPLYFVSFVLTAQFVLINVVVAVLMKHLDDSNKEA
QEDAEMDAELELEMAHGLGPGPRLPTGSPGAPGRGPGGAGGGGDTEGGLCRRCYSPAQEN
LWLDSVSLIIKDSLEGELTIIDNLSGSIFHHYSSPAGCKKCHHDKQEVQLAETEAFSLNS
DRSSSILLGDDLSLEDPTACPPGRKDSKGELDPPEPMRVGDLGECFFPLSSTAVSPDPEN
FLCEMEEIPFNPVRSWLKHDSSQAPPSPFSPDASSPLLPMPAEFFHPAVSASQKGPEKGT
GTGTLPKIALQGSWASLRSPRVNCTLLRQATGSDTSLDASPSSSAGSLQTTLEDSLTLSD
SPRRALGPPAPAPGPRAGLSPAARRRLSLRGRGLFSLRGLRAHQRSHSSGGSTSPGCTHH
DSMDPSDEEGRGGAGGGGAGSEHSETLSSLSLTSLFCPPPPPPAPGLTPARKFSSTSSLA
APGRPHAAALAHGLARSPSWAADRSKDPPGRAPLPMGLGPLAPPPQPLPGELEPGDAASK
RKR
|
|
|
|---|
| BDBM50240424 |
|---|
| n/a |
|---|
| Name | BDBM50240424 |
|---|
| Synonyms: | 3-Ethyl-3-methyl-pyrrolidine-2,5-dione | 3-Ethyl-3-methyl-pyrrolidine-2,5-dione (ethosuximide) | 3-Ethyl-3-methyl-pyrrolidine-2,5-dione(ethosuximide) | 3-ethyl-3-methylpyrrolidine-2,5-dione | CHEMBL696 | CI-366 | CN-10395 | ETHOSUXIMIDE | PM-671 | Zarontin | zarontin3-Ethyl-3-methyl-pyrrolidine-2,5-dione |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C7H11NO2 |
|---|
| Mol. Mass. | 141.1677 |
|---|
| SMILES | CCC1(C)CC(=O)NC1=O |
|---|
| Structure | 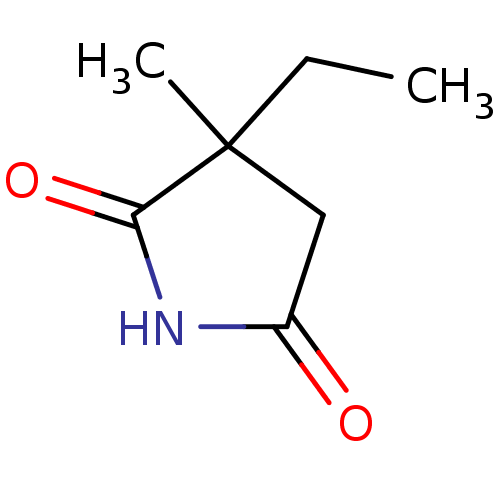
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Yang, ZQ; Barrow, JC; Shipe, WD; Schlegel, KA; Shu, Y; Yang, FV; Lindsley, CW; Rittle, KE; Bock, MG; Hartman, GD; Uebele, VN; Nuss, CE; Fox, SV; Kraus, RL; Doran, SM; Connolly, TM; Tang, C; Ballard, JE; Kuo, Y; Adarayan, ED; Prueksaritanont, T; Zrada, MM; Marino, MJ; Graufelds, VK; DiLella, AG; Reynolds, IJ; Vargas, HM; Bunting, PB; Woltmann, RF; Magee, MM; Koblan, KS; Renger, JJ Discovery of 1,4-substituted piperidines as potent and selective inhibitors of T-type calcium channels. J Med Chem51:6471-7 (2008) [PubMed] Article
Yang, ZQ; Barrow, JC; Shipe, WD; Schlegel, KA; Shu, Y; Yang, FV; Lindsley, CW; Rittle, KE; Bock, MG; Hartman, GD; Uebele, VN; Nuss, CE; Fox, SV; Kraus, RL; Doran, SM; Connolly, TM; Tang, C; Ballard, JE; Kuo, Y; Adarayan, ED; Prueksaritanont, T; Zrada, MM; Marino, MJ; Graufelds, VK; DiLella, AG; Reynolds, IJ; Vargas, HM; Bunting, PB; Woltmann, RF; Magee, MM; Koblan, KS; Renger, JJ Discovery of 1,4-substituted piperidines as potent and selective inhibitors of T-type calcium channels. J Med Chem51:6471-7 (2008) [PubMed] Article