| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Stromelysin-1 |
|---|
| Ligand | BDBM50269001 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_523766 (CHEMBL1008374) |
|---|
| IC50 | >5000±n/a nM |
|---|
| Citation |  Burns, DM; Li, YL; Shi, E; He, C; Xu, M; Zhuo, J; Zhang, C; Qian, DQ; Li, Y; Wynn, R; Covington, MB; Katiyar, K; Marando, CA; Fridman, JS; Scherle, P; Friedman, S; Metcalf, B; Yao, W Compelling P1 substituent affect on metalloprotease binding profile enables the design of a novel cyclohexyl core scaffold with excellent MMP selectivity and HER-2 sheddase inhibition. Bioorg Med Chem Lett19:3525-30 (2009) [PubMed] Article Burns, DM; Li, YL; Shi, E; He, C; Xu, M; Zhuo, J; Zhang, C; Qian, DQ; Li, Y; Wynn, R; Covington, MB; Katiyar, K; Marando, CA; Fridman, JS; Scherle, P; Friedman, S; Metcalf, B; Yao, W Compelling P1 substituent affect on metalloprotease binding profile enables the design of a novel cyclohexyl core scaffold with excellent MMP selectivity and HER-2 sheddase inhibition. Bioorg Med Chem Lett19:3525-30 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Stromelysin-1 |
|---|
| Name: | Stromelysin-1 |
|---|
| Synonyms: | MMP-3 | MMP3 | MMP3_HUMAN | Matrix metalloproteinase (2 and 3) | Matrix metalloproteinase 3 | Matrix metalloproteinase-3 | Matrix metalloproteinase-3 (MMP-3) | Matrix metalloproteinase-3 (MMP3) | SL-1 | STMY1 | Stromelysin 1 | Transin-1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 53973.13 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P08254 |
|---|
| Residue: | 477 |
|---|
| Sequence: | MKSLPILLLLCVAVCSAYPLDGAARGEDTSMNLVQKYLENYYDLKKDVKQFVRRKDSGPV
VKKIREMQKFLGLEVTGKLDSDTLEVMRKPRCGVPDVGHFRTFPGIPKWRKTHLTYRIVN
YTPDLPKDAVDSAVEKALKVWEEVTPLTFSRLYEGEADIMISFAVREHGDFYPFDGPGNV
LAHAYAPGPGINGDAHFDDDEQWTKDTTGTNLFLVAAHEIGHSLGLFHSANTEALMYPLY
HSLTDLTRFRLSQDDINGIQSLYGPPPDSPETPLVPTEPVPPEPGTPANCDPALSFDAVS
TLRGEILIFKDRHFWRKSLRKLEPELHLISSFWPSLPSGVDAAYEVTSKDLVFIFKGNQF
WAIRGNEVRAGYPRGIHTLGFPPTVRKIDAAISDKEKNKTYFFVEDKYWRFDEKRNSMEP
GFPKQIAEDFPGIDSKIDAVFEEFGFFYFFTGSSQLEFDPNAKKVTHTLKSNSWLNC
|
|
|
|---|
| BDBM50269001 |
|---|
| n/a |
|---|
| Name | BDBM50269001 |
|---|
| Synonyms: | (1S,2S,5RS)-N-hydroxy-5-(2-(4-methylpiperazin-1-yl)-2-oxoethyl)-2-(4-phenylpiperazine-1-carbonyl)cyclohexanecarboxamide | CHEMBL497222 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H37N5O4 |
|---|
| Mol. Mass. | 471.5924 |
|---|
| SMILES | CN1CCN(CC1)C(=O)CC1CC[C@@H]([C@H](C1)C(=O)NO)C(=O)N1CCN(CC1)c1ccccc1 |r| |
|---|
| Structure | 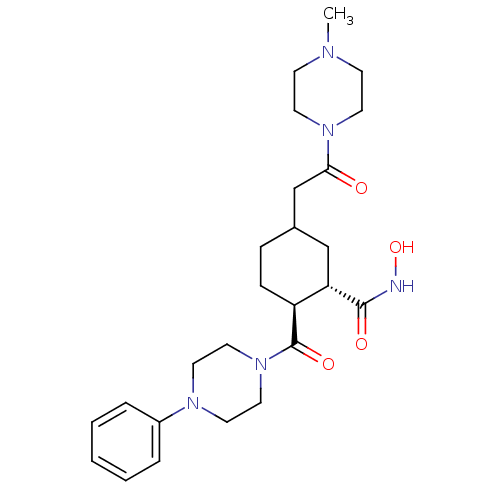
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Burns, DM; Li, YL; Shi, E; He, C; Xu, M; Zhuo, J; Zhang, C; Qian, DQ; Li, Y; Wynn, R; Covington, MB; Katiyar, K; Marando, CA; Fridman, JS; Scherle, P; Friedman, S; Metcalf, B; Yao, W Compelling P1 substituent affect on metalloprotease binding profile enables the design of a novel cyclohexyl core scaffold with excellent MMP selectivity and HER-2 sheddase inhibition. Bioorg Med Chem Lett19:3525-30 (2009) [PubMed] Article
Burns, DM; Li, YL; Shi, E; He, C; Xu, M; Zhuo, J; Zhang, C; Qian, DQ; Li, Y; Wynn, R; Covington, MB; Katiyar, K; Marando, CA; Fridman, JS; Scherle, P; Friedman, S; Metcalf, B; Yao, W Compelling P1 substituent affect on metalloprotease binding profile enables the design of a novel cyclohexyl core scaffold with excellent MMP selectivity and HER-2 sheddase inhibition. Bioorg Med Chem Lett19:3525-30 (2009) [PubMed] Article