| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Alpha-1A/Alpha-1B/Alpha-1D adrenergic receptor |
|---|
| Ligand | BDBM50289383 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_32554 (CHEMBL645689) |
|---|
| Ki | 8±n/a nM |
|---|
| Citation |  Perrone, R; Berardi, F; Colabufo, NA; Leopoldo, M; Tortorella, V 1-(2-METHOXYPHENYL)-4-ALKYLPIPERAZINES: EFFECT OF THE N-4 SUBSTITUENT ON THE AFFINITY AND SELECTIVITY FOR DOPAMINE D4 RECEPTOR Bioorg Med Chem Lett7:1327-1330 (1997) Article Perrone, R; Berardi, F; Colabufo, NA; Leopoldo, M; Tortorella, V 1-(2-METHOXYPHENYL)-4-ALKYLPIPERAZINES: EFFECT OF THE N-4 SUBSTITUENT ON THE AFFINITY AND SELECTIVITY FOR DOPAMINE D4 RECEPTOR Bioorg Med Chem Lett7:1327-1330 (1997) Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Alpha-1A/Alpha-1B/Alpha-1D adrenergic receptor |
|---|
| Name: | Alpha-1A/Alpha-1B/Alpha-1D adrenergic receptor |
|---|
| Synonyms: | Adrenergic receptor alpha-1 | Alpha-1 Adrenergic Receptor/ adrenergic receptor/ adrenergic receptor | alpha-1 Adrenergic Receptor |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of EBI is 303470 |
|---|
| Components: | This complex has 3 components. |
|---|
| Component 1 |
| Name: | Alpha-1B adrenergic receptor |
|---|
| Synonyms: | ADA1B_RAT | Adra1b | Alpha 1B-adrenoceptor | Alpha 1B-adrenoreceptor | Alpha adrenergic receptor 1A and 1B | Alpha-1 Adrenergic Receptor | Alpha-1Adrenoceptor | Alpha-1B adrenergic receptor | Alpha-1B adrenoreceptor | adrenergic Alpha1B |
|---|
| Type: | G Protein-Coupled Receptor (GPCR) |
|---|
| Mol. Mass.: | 56606.71 |
|---|
| Organism: | Rattus norvegicus (rat) |
|---|
| Description: | Receptor binding assays were performed using rat cortical membranes. |
|---|
| Residue: | 515 |
|---|
| Sequence: | MNPDLDTGHNTSAPAHWGELKDDNFTGPNQTSSNSTLPQLDVTRAISVGLVLGAFILFAI
VGNILVILSVACNRHLRTPTNYFIVNLAIADLLLSFTVLPFSATLEVLGYWVLGRIFCDI
WAAVDVLCCTASILSLCAISIDRYIGVRYSLQYPTLVTRRKAILALLSVWVLSTVISIGP
LLGWKEPAPNDDKECGVTEEPFYALFSSLGSFYIPLAVILVMYCRVYIVAKRTTKNLEAG
VMKEMSNSKELTLRIHSKNFHEDTLSSTKAKGHNPRSSIAVKLFKFSREKKAAKTLGIVV
GMFILCWLPFFIALPLGSLFSTLKPPDAVFKVVFWLGYFNSCLNPIIYPCSSKEFKRAFM
RILGCQCRGGRRRRRRRRLGACAYTYRPWTRGGSLERSQSRKDSLDDSGSCMSGTQRTLP
SASPSPGYLGRGTQPPVELCAFPEWKPGALLSLPEPPGRRGRLDSGPLFTFKLLGDPESP
GTEGDTSNGGCDTTTDLANGQPGFKSNMPLAPGHF
|
|
|
|---|
| Component 2 |
| Name: | Alpha-1A adrenergic receptor |
|---|
| Synonyms: | ADA1A_RAT | Adra1a | Adra1c | Alpha-1 adrenoreceptor | Alpha-1A adrenoceptor | Alpha-1A adrenoreceptor | Alpha-1C adrenergic receptor | adrenergic Alpha1A |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 51620.15 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | P43140 |
|---|
| Residue: | 466 |
|---|
| Sequence: | MVLLSENASEGSNCTHPPAPVNISKAILLGVILGGLIIFGVLGNILVILSVACHRHLHSV
THYYIVNLAVADLLLTSTVLPFSAIFEILGYWAFGRVFCNIWAAVDVLCCTASIMGLCII
SIDRYIGVSYPLRYPTIVTQRRGVRALLCVWVLSLVISIGPLFGWRQPAPEDETICQINE
EPGYVLFSALGSFYVPLAIILVMYCRVYVVAKRESRGLKSGLKTDKSDSEQVTLRIHRKN
VPAEGGGVSSAKNKTHFSVRLLKFSREKKAAKTLGIVVGCFVLCWLPFFLVMPIGSFFPD
FKPSETVFKIVFWLGYLNSCINPIIYPCSSQEFKKAFQNVLRIQCLRRRQSSKHALGYTL
HPPSQALEGQHRDMVRIPVGSGETFYKISKTDGVCEWKFFSSMPQGSARITVPKDQSACT
TARVRSKSFLQVCCCVGSSAPRPEENHQVPTIKIHTISLGENGEEV
|
|
|
|---|
| Component 3 |
| Name: | Alpha-1D adrenergic receptor |
|---|
| Synonyms: | ADA1D_RAT | Adra1a | Adra1d | Alpha adrenergic receptor 1A and 1D | Alpha-1D adrenergic receptor | Serotonin 1a (5-HT1a) receptor/Adrenergic receptor alpha-1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 59375.97 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | P23944 |
|---|
| Residue: | 561 |
|---|
| Sequence: | MTFRDILSVTFEGPRSSSSTGGSGAGGGAGTVGPEGGAVGGVPGATGGGAVVGTGSGEDN
QSSTGEPGAAASGEVNGSAAVGGLVVSAQGVGVGVFLAAFILTAVAGNLLVILSVACNRH
LQTVTNYFIVNLAVADLLLSAAVLPFSATMEVLGFWAFGRTFCDVWAAVDVLCCTASILS
LCTISVDRYVGVRHSLKYPAIMTERKAAAILALLWAVALVVSVGPLLGWKEPVPPDERFC
GITEEVGYAIFSSVCSFYLPMAVIVVMYCRVYVVARSTTRSLEAGIKREPGKASEVVLRI
HCRGAATSAKGYPGTQSSKGHTLRSSLSVRLLKFSREKKAAKTLAIVVGVFVLCWFPFFF
VLPLGSLFPQLKPSEGVFKVIFWLGYFNSCVNPLIYPCSSREFKRAFLRLLRCQCRRRRR
RLWAVYGHHWRASTGDARSDCAPSPRIAPPGAPLALTAHPGAGSADTPETQDSVSSSRKP
ASALREWRLLGPLQRPTTQLRAKVSSLSHKIRSGARRAETACALRSEVEAVSLNVPQDGA
EAVICQAYEPGDYSNLRETDI
|
|
|
|---|
| BDBM50289383 |
|---|
| n/a |
|---|
| Name | BDBM50289383 |
|---|
| Synonyms: | 4-Amino-5-chloro-2-methoxy-N-{2-[4-(2-methoxy-phenyl)-piperazin-1-yl]-ethyl}-benzamide | CHEMBL26758 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H27ClN4O3 |
|---|
| Mol. Mass. | 418.917 |
|---|
| SMILES | COc1ccccc1N1CCN(CCNC(=O)c2cc(Cl)c(N)cc2OC)CC1 |
|---|
| Structure | 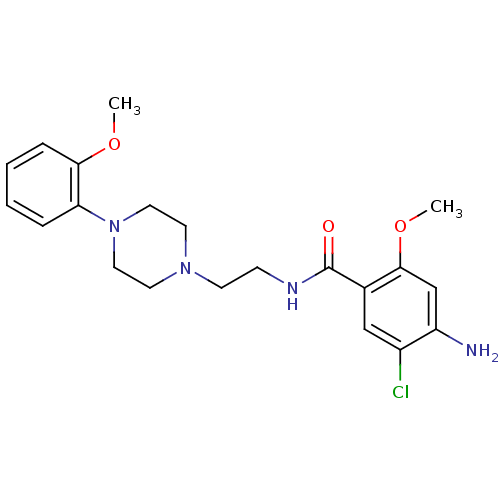
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Perrone, R; Berardi, F; Colabufo, NA; Leopoldo, M; Tortorella, V 1-(2-METHOXYPHENYL)-4-ALKYLPIPERAZINES: EFFECT OF THE N-4 SUBSTITUENT ON THE AFFINITY AND SELECTIVITY FOR DOPAMINE D4 RECEPTOR Bioorg Med Chem Lett7:1327-1330 (1997) Article
Perrone, R; Berardi, F; Colabufo, NA; Leopoldo, M; Tortorella, V 1-(2-METHOXYPHENYL)-4-ALKYLPIPERAZINES: EFFECT OF THE N-4 SUBSTITUENT ON THE AFFINITY AND SELECTIVITY FOR DOPAMINE D4 RECEPTOR Bioorg Med Chem Lett7:1327-1330 (1997) Article