| Reaction Details |
|---|
 | Report a problem with these data |
| Target | NAD-dependent protein deacetylase sirtuin-1 |
|---|
| Ligand | BDBM50228364 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_495772 (CHEMBL1007891) |
|---|
| Kd | 16200±n/a nM |
|---|
| Citation |  Vu, CB; Bemis, JE; Disch, JS; Ng, PY; Nunes, JJ; Milne, JC; Carney, DP; Lynch, AV; Smith, JJ; Lavu, S; Lambert, PD; Gagne, DJ; Jirousek, MR; Schenk, S; Olefsky, JM; Perni, RB Discovery of imidazo[1,2-b]thiazole derivatives as novel SIRT1 activators. J Med Chem52:1275-83 (2010) [PubMed] Article Vu, CB; Bemis, JE; Disch, JS; Ng, PY; Nunes, JJ; Milne, JC; Carney, DP; Lynch, AV; Smith, JJ; Lavu, S; Lambert, PD; Gagne, DJ; Jirousek, MR; Schenk, S; Olefsky, JM; Perni, RB Discovery of imidazo[1,2-b]thiazole derivatives as novel SIRT1 activators. J Med Chem52:1275-83 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| NAD-dependent protein deacetylase sirtuin-1 |
|---|
| Name: | NAD-dependent protein deacetylase sirtuin-1 |
|---|
| Synonyms: | NAD-Dependent Deacetylase Sirtuin-1 | NAD-dependent deacetylase sirtuin 1 | NAD-dependent protein deacetylase sirtuin-1 (SIRT1) | SIR1_HUMAN | SIR2-like protein 1 | SIR2L1 | SIRT1 | Sirtuin 1 (SIRT1) | Sirtuin-1 (SIRT1) | hSIR2 |
|---|
| Type: | Developmental protein; hydrolase |
|---|
| Mol. Mass.: | 81626.66 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 747 |
|---|
| Sequence: | MADEAALALQPGGSPSAAGADREAASSPAGEPLRKRPRRDGPGLERSPGEPGGAAPEREV
PAAARGCPGAAAAALWREAEAEAAAAGGEQEAQATAAAGEGDNGPGLQGPSREPPLADNL
YDEDDDDEGEEEEEAAAAAIGYRDNLLFGDEIITNGFHSCESDEEDRASHASSSDWTPRP
RIGPYTFVQQHLMIGTDPRTILKDLLPETIPPPELDDMTLWQIVINILSEPPKRKKRKDI
NTIEDAVKLLQECKKIIVLTGAGVSVSCGIPDFRSRDGIYARLAVDFPDLPDPQAMFDIE
YFRKDPRPFFKFAKEIYPGQFQPSLCHKFIALSDKEGKLLRNYTQNIDTLEQVAGIQRII
QCHGSFATASCLICKYKVDCEAVRGDIFNQVVPRCPRCPADEPLAIMKPEIVFFGENLPE
QFHRAMKYDKDEVDLLIVIGSSLKVRPVALIPSSIPHEVPQILINREPLPHLHFDVELLG
DCDVIINELCHRLGGEYAKLCCNPVKLSEITEKPPRTQKELAYLSELPPTPLHVSEDSSS
PERTSPPDSSVIVTLLDQAAKSNDDLDVSESKGCMEEKPQEVQTSRNVESIAEQMENPDL
KNVGSSTGEKNERTSVAGTVRKCWPNRVAKEQISRRLDGNQYLFLPPNRYIFHGAEVYSD
SEDDVLSSSSCGSNSDSGTCQSPSLEEPMEDESEIEEFYNGLEDEPDVPERAGGAGFGTD
GDDQEAINEAISVKQEVTDMNYPSNKS
|
|
|
|---|
| BDBM50228364 |
|---|
| n/a |
|---|
| Name | BDBM50228364 |
|---|
| Synonyms: | 3,4,5-trimethoxy-N-(2-(3-(piperazin-1-ylmethyl)imidazo[2,1-b]thiazol-6-yl)phenyl)benzamide | CHEMBL254156 | SRT-1460 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H29N5O4S |
|---|
| Mol. Mass. | 507.605 |
|---|
| SMILES | COc1cc(cc(OC)c1OC)C(=O)Nc1ccccc1-c1cn2c(CN3CCNCC3)csc2n1 |
|---|
| Structure | 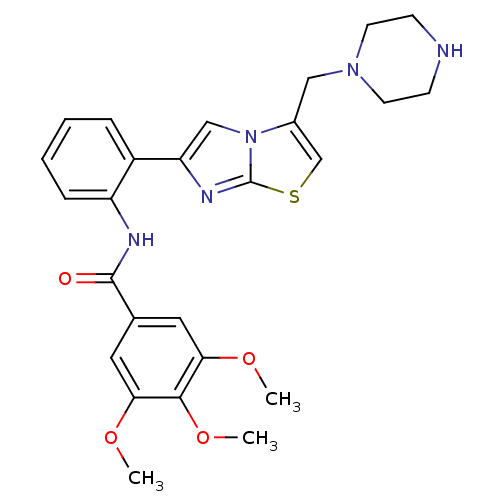
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Vu, CB; Bemis, JE; Disch, JS; Ng, PY; Nunes, JJ; Milne, JC; Carney, DP; Lynch, AV; Smith, JJ; Lavu, S; Lambert, PD; Gagne, DJ; Jirousek, MR; Schenk, S; Olefsky, JM; Perni, RB Discovery of imidazo[1,2-b]thiazole derivatives as novel SIRT1 activators. J Med Chem52:1275-83 (2010) [PubMed] Article
Vu, CB; Bemis, JE; Disch, JS; Ng, PY; Nunes, JJ; Milne, JC; Carney, DP; Lynch, AV; Smith, JJ; Lavu, S; Lambert, PD; Gagne, DJ; Jirousek, MR; Schenk, S; Olefsky, JM; Perni, RB Discovery of imidazo[1,2-b]thiazole derivatives as novel SIRT1 activators. J Med Chem52:1275-83 (2010) [PubMed] Article