| Citation |  Schnute, ME; O'Brien, PM; Nahra, J; Morris, M; Howard Roark, W; Hanau, CE; Ruminski, PG; Scholten, JA; Fletcher, TR; Hamper, BC; Carroll, JN; Patt, WC; Shieh, HS; Collins, B; Pavlovsky, AG; Palmquist, KE; Aston, KW; Hitchcock, J; Rogers, MD; McDonald, J; Johnson, AR; Munie, GE; Wittwer, AJ; Man, CF; Settle, SL; Nemirovskiy, O; Vickery, LE; Agawal, A; Dyer, RD; Sunyer, T Discovery of (pyridin-4-yl)-2H-tetrazole as a novel scaffold to identify highly selective matrix metalloproteinase-13 inhibitors for the treatment of osteoarthritis. Bioorg Med Chem Lett20:576-80 (2010) [PubMed] Article Schnute, ME; O'Brien, PM; Nahra, J; Morris, M; Howard Roark, W; Hanau, CE; Ruminski, PG; Scholten, JA; Fletcher, TR; Hamper, BC; Carroll, JN; Patt, WC; Shieh, HS; Collins, B; Pavlovsky, AG; Palmquist, KE; Aston, KW; Hitchcock, J; Rogers, MD; McDonald, J; Johnson, AR; Munie, GE; Wittwer, AJ; Man, CF; Settle, SL; Nemirovskiy, O; Vickery, LE; Agawal, A; Dyer, RD; Sunyer, T Discovery of (pyridin-4-yl)-2H-tetrazole as a novel scaffold to identify highly selective matrix metalloproteinase-13 inhibitors for the treatment of osteoarthritis. Bioorg Med Chem Lett20:576-80 (2010) [PubMed] Article |
|---|
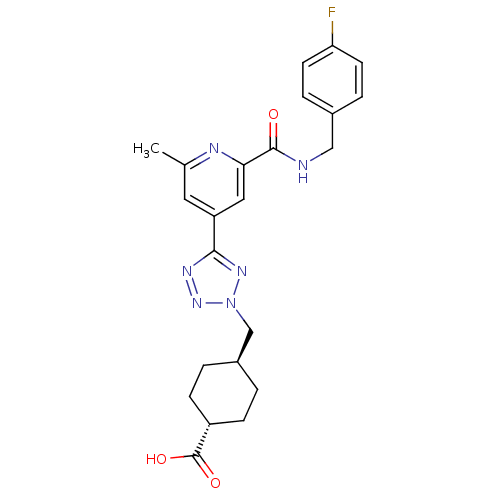
| SMILES | Cc1cc(cc(n1)C(=O)NCc1ccc(F)cc1)-c1nnn(C[C@H]2CC[C@@H](CC2)C(O)=O)n1 |r,wU:26.31,wD:23.24,(30.11,-12.24,;29.32,-10.92,;30.07,-9.57,;29.28,-8.26,;27.74,-8.27,;26.98,-9.62,;27.77,-10.94,;25.44,-9.64,;24.69,-10.98,;24.66,-8.31,;23.12,-8.33,;22.33,-7.01,;23.08,-5.67,;22.3,-4.34,;20.75,-4.36,;19.97,-3.04,;20,-5.72,;20.79,-7.03,;30.03,-6.91,;29.39,-5.51,;30.52,-4.46,;31.87,-5.21,;33.27,-4.57,;34.61,-5.32,;34.63,-6.85,;35.98,-7.6,;37.3,-6.81,;37.27,-5.26,;35.93,-4.52,;38.65,-7.56,;38.68,-9.1,;39.97,-6.77,;31.57,-6.73,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Schnute, ME; O'Brien, PM; Nahra, J; Morris, M; Howard Roark, W; Hanau, CE; Ruminski, PG; Scholten, JA; Fletcher, TR; Hamper, BC; Carroll, JN; Patt, WC; Shieh, HS; Collins, B; Pavlovsky, AG; Palmquist, KE; Aston, KW; Hitchcock, J; Rogers, MD; McDonald, J; Johnson, AR; Munie, GE; Wittwer, AJ; Man, CF; Settle, SL; Nemirovskiy, O; Vickery, LE; Agawal, A; Dyer, RD; Sunyer, T Discovery of (pyridin-4-yl)-2H-tetrazole as a novel scaffold to identify highly selective matrix metalloproteinase-13 inhibitors for the treatment of osteoarthritis. Bioorg Med Chem Lett20:576-80 (2010) [PubMed] Article
Schnute, ME; O'Brien, PM; Nahra, J; Morris, M; Howard Roark, W; Hanau, CE; Ruminski, PG; Scholten, JA; Fletcher, TR; Hamper, BC; Carroll, JN; Patt, WC; Shieh, HS; Collins, B; Pavlovsky, AG; Palmquist, KE; Aston, KW; Hitchcock, J; Rogers, MD; McDonald, J; Johnson, AR; Munie, GE; Wittwer, AJ; Man, CF; Settle, SL; Nemirovskiy, O; Vickery, LE; Agawal, A; Dyer, RD; Sunyer, T Discovery of (pyridin-4-yl)-2H-tetrazole as a novel scaffold to identify highly selective matrix metalloproteinase-13 inhibitors for the treatment of osteoarthritis. Bioorg Med Chem Lett20:576-80 (2010) [PubMed] Article