

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Mu-type opioid receptor | ||
| Ligand | BDBM50311489 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_622958 (CHEMBL1112210) | ||
| Ki | 346±n/a nM | ||
| Citation |  Palin, R; Clark, JK; Evans, L; Feilden, H; Fletcher, D; Hamilton, NM; Houghton, AK; Jones, PS; McArthur, D; Montgomery, B; Ratcliffe, PD; Smith, AR; Sutherland, A; Weston, MA; Wishart, G Rapid access towards follow-up NOP receptor agonists using a knowledge based approach. Bioorg Med Chem Lett19:6441-6 (2009) [PubMed] Article Palin, R; Clark, JK; Evans, L; Feilden, H; Fletcher, D; Hamilton, NM; Houghton, AK; Jones, PS; McArthur, D; Montgomery, B; Ratcliffe, PD; Smith, AR; Sutherland, A; Weston, MA; Wishart, G Rapid access towards follow-up NOP receptor agonists using a knowledge based approach. Bioorg Med Chem Lett19:6441-6 (2009) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Mu-type opioid receptor | |||
| Name: | Mu-type opioid receptor | ||
| Synonyms: | M-OR-1 | MOP | MOR-1 | MOR1 | MUOR1 | Mu Opioid Receptor | Mu opiate receptor | OPIATE Mu | OPRM1 | OPRM_HUMAN | hMOP | mu-type opioid receptor isoform MOR-1 | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 44789.51 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P35372 | ||
| Residue: | 400 | ||
| Sequence: |
| ||
| BDBM50311489 | |||
| n/a | |||
| Name | BDBM50311489 | ||
| Synonyms: | 2-{3-[1-(4-Isopropyl-cyclohexyl)-piperidin-4-yl]-2,2-dioxo-2,3-dihydro-2lambda*6*-benzo[1,2,5]thiadiazol-1-yl}-N-methyl-acetamide | CHEMBL1076242 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H36N4O3S | ||
| Mol. Mass. | 448.622 | ||
| SMILES | CNC(=O)CN1c2ccccc2N(C2CCN(CC2)C2CCC(CC2)C(C)C)S1(=O)=O |(3.05,-10.56,;1.51,-10.56,;.74,-11.9,;1.51,-13.23,;-.8,-11.9,;-1.57,-13.23,;-1.24,-14.73,;.1,-15.49,;.11,-17.04,;-1.23,-17.81,;-2.56,-17.04,;-2.56,-15.5,;-3.7,-14.48,;-5.03,-15.26,;-6.36,-14.5,;-7.69,-15.27,;-7.69,-16.81,;-6.35,-17.58,;-5.01,-16.8,;-9.02,-17.59,;-10.35,-16.82,;-11.68,-17.59,;-11.68,-19.13,;-10.35,-19.9,;-9.01,-19.13,;-13.02,-19.89,;-14.35,-19.12,;-13.03,-21.43,;-3.09,-13.08,;-2.7,-11.58,;-4.43,-12.31,)| | ||
| Structure | 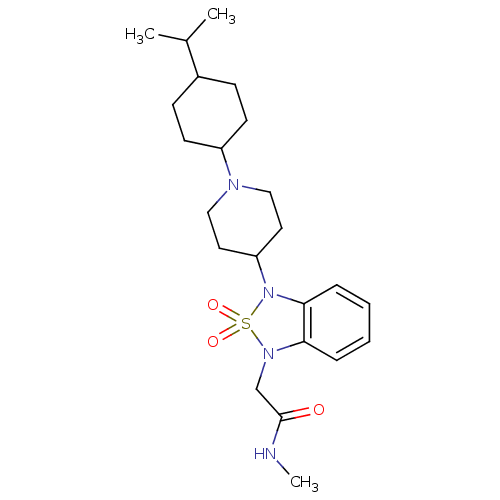
| ||