| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Sucrase-isomaltase, intestinal |
|---|
| Ligand | BDBM50312527 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_617530 (CHEMBL1099880) |
|---|
| IC50 | >1000000±n/a nM |
|---|
| Citation |  Wennekes, T; Meijer, AJ; Groen, AK; Boot, RG; Groener, JE; van Eijk, M; Ottenhoff, R; Bijl, N; Ghauharali, K; Song, H; O'Shea, TJ; Liu, H; Yew, N; Copeland, D; van den Berg, RJ; van der Marel, GA; Overkleeft, HS; Aerts, JM Dual-action lipophilic iminosugar improves glycemic control in obese rodents by reduction of visceral glycosphingolipids and buffering of carbohydrate assimilation. J Med Chem53:689-98 (2010) [PubMed] Article Wennekes, T; Meijer, AJ; Groen, AK; Boot, RG; Groener, JE; van Eijk, M; Ottenhoff, R; Bijl, N; Ghauharali, K; Song, H; O'Shea, TJ; Liu, H; Yew, N; Copeland, D; van den Berg, RJ; van der Marel, GA; Overkleeft, HS; Aerts, JM Dual-action lipophilic iminosugar improves glycemic control in obese rodents by reduction of visceral glycosphingolipids and buffering of carbohydrate assimilation. J Med Chem53:689-98 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Sucrase-isomaltase, intestinal |
|---|
| Name: | Sucrase-isomaltase, intestinal |
|---|
| Synonyms: | Alpha glucosidase | Isomaltase | SI | SUIS_HUMAN | Sucrase | Sucrase-isomaltase | Sucrase-isomaltase, intestinal |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 209423.23 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1435477 |
|---|
| Residue: | 1827 |
|---|
| Sequence: | MARKKFSGLEISLIVLFVIVTIIAIALIVVLATKTPAVDEISDSTSTPATTRVTTNPSDS
GKCPNVLNDPVNVRINCIPEQFPTEGICAQRGCCWRPWNDSLIPWCFFVDNHGYNVQDMT
TTSIGVEAKLNRIPSPTLFGNDINSVLFTTQNQTPNRFRFKITDPNNRRYEVPHQYVKEF
TGPTVSDTLYDVKVAQNPFSIQVIRKSNGKTLFDTSIGPLVYSDQYLQISTRLPSDYIYG
IGEQVHKRFRHDLSWKTWPIFTRDQLPGDNNNNLYGHQTFFMCIEDTSGKSFGVFLMNSN
AMEIFIQPTPIVTYRVTGGILDFYILLGDTPEQVVQQYQQLVGLPAMPAYWNLGFQLSRW
NYKSLDVVKEVVRRNREAGIPFDTQVTDIDYMEDKKDFTYDQVAFNGLPQFVQDLHDHGQ
KYVIILDPAISIGRRANGTTYATYERGNTQHVWINESDGSTPIIGEVWPGLTVYPDFTNP
NCIDWWANECSIFHQEVQYDGLWIDMNEVSSFIQGSTKGCNVNKLNYPPFTPDILDKLMY
SKTICMDAVQNWGKQYDVHSLYGYSMAIATEQAVQKVFPNKRSFILTRSTFAGSGRHAAH
WLGDNTASWEQMEWSITGMLEFSLFGIPLVGADICGFVAETTEELCRRWMQLGAFYPFSR
NHNSDGYEHQDPAFFGQNSLLVKSSRQYLTIRYTLLPFLYTLFYKAHVFGETVARPVLHE
FYEDTNSWIEDTEFLWGPALLITPVLKQGADTVSAYIPDAIWYDYESGAKRPWRKQRVDM
YLPADKIGLHLRGGYIIPIQEPDVTTTASRKNPLGLIVALGENNTAKGDFFWDDGETKDT
IQNGNYILYTFSVSNNTLDIVCTHSSYQEGTTLAFQTVKILGLTDSVTEVRVAENNQPMN
AHSNFTYDASNQVLLIADLKLNLGRNFSVQWNQIFSENERFNCYPDADLATEQKCTQRGC
VWRTGSSLSKAPECYFPRQDNSYSVNSARYSSMGITADLQLNTANARIKLPSDPISTLRV
EVKYHKNDMLQFKIYDPQKKRYEVPVPLNIPTTPISTYEDRLYDVEIKENPFGIQIRRRS
SGRVIWDSWLPGFAFNDQFIQISTRLPSEYIYGFGEVEHTAFKRDLNWNTWGMFTRDQPP
GYKLNSYGFHPYYMALEEEGNAHGVFLLNSNAMDVTFQPTPALTYRTVGGILDFYMFLGP
TPEVATKQYHEVIGHPVMPAYWALGFQLCRYGYANTSEVRELYDAMVAANIPYDVQYTDI
DYMERQLDFTIGEAFQDLPQFVDKIRGEGMRYIIILDPAISGNETKTYPAFERGQQNDVF
VKWPNTNDICWAKVWPDLPNITIDKTLTEDEAVNASRAHVAFPDFFRTSTAEWWAREIVD
FYNEKMKFDGLWIDMNEPSSFVNGTTTNQCRNDELNYPPYFPELTKRTDGLHFRTICMEA
EQILSDGTSVLHYDVHNLYGWSQMKPTHDALQKTTGKRGIVISRSTYPTSGRWGGHWLGD
NYARWDNMDKSIIGMMEFSLFGMSYTGADICGFFNNSEYHLCTRWMQLGAFYPYSRNHNI
ANTRRQDPASWNETFAEMSRNILNIRYTLLPYFYTQMHEIHANGGTVIRPLLHEFFDEKP
TWDIFKQFLWGPAFMVTPVLEPYVQTVNAYVPNARWFDYHTGKDIGVRGQFQTFNASYDT
INLHVRGGHILPCQEPAQNTFYSRQKHMKLIVAADDNQMAQGSLFWDDGESIDTYERDLY
LSVQFNLNQTTLTSTILKRGYINKSETRLGSLHVWGKGTTPVNAVTLTYNGNKNSLPFNE
DTTNMILRIDLTTHNVTLEEPIEINWS
|
|
|
|---|
| BDBM50312527 |
|---|
| n/a |
|---|
| Name | BDBM50312527 |
|---|
| Synonyms: | CHEMBL1086996 | N-[5-(Adamantan-1-yl-methoxy)-pentyl]-L-ido-1-deoxynojirimycin |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H39NO5 |
|---|
| Mol. Mass. | 397.5488 |
|---|
| SMILES | OC[C@H]1[C@@H](O)[C@H](O)[C@@H](O)CN1CCCCCOCC12CC3CC(CC(C3)C1)C2 |r,TLB:21:22:20.25.19:26,THB:21:20:22.23.27:26,23:22:19:25.24.26,23:24:21.22.27:19| |
|---|
| Structure | 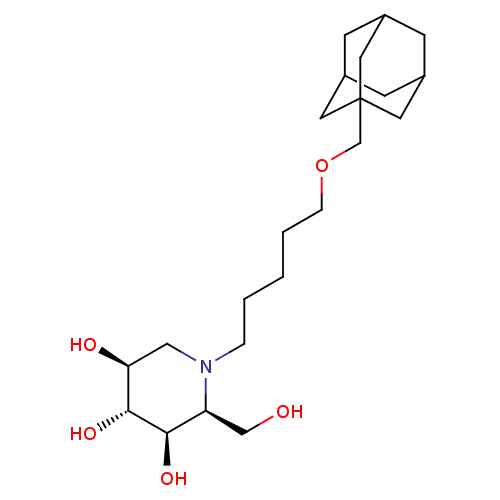
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wennekes, T; Meijer, AJ; Groen, AK; Boot, RG; Groener, JE; van Eijk, M; Ottenhoff, R; Bijl, N; Ghauharali, K; Song, H; O'Shea, TJ; Liu, H; Yew, N; Copeland, D; van den Berg, RJ; van der Marel, GA; Overkleeft, HS; Aerts, JM Dual-action lipophilic iminosugar improves glycemic control in obese rodents by reduction of visceral glycosphingolipids and buffering of carbohydrate assimilation. J Med Chem53:689-98 (2010) [PubMed] Article
Wennekes, T; Meijer, AJ; Groen, AK; Boot, RG; Groener, JE; van Eijk, M; Ottenhoff, R; Bijl, N; Ghauharali, K; Song, H; O'Shea, TJ; Liu, H; Yew, N; Copeland, D; van den Berg, RJ; van der Marel, GA; Overkleeft, HS; Aerts, JM Dual-action lipophilic iminosugar improves glycemic control in obese rodents by reduction of visceral glycosphingolipids and buffering of carbohydrate assimilation. J Med Chem53:689-98 (2010) [PubMed] Article