| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 3 |
|---|
| Ligand | BDBM50314136 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_623893 (CHEMBL1105416) |
|---|
| IC50 | 58±n/a nM |
|---|
| Citation |  Oger, F; Lecorgne, A; Sala, E; Nardese, V; Demay, F; Chevance, S; Desravines, DC; Aleksandrova, N; Le Guével, R; Lorenzi, S; Beccari, AR; Barath, P; Hart, DJ; Bondon, A; Carettoni, D; Simonneaux, G; Salbert, G Biological and biophysical properties of the histone deacetylase inhibitor suberoylanilide hydroxamic acid are affected by the presence of short alkyl groups on the phenyl ring. J Med Chem53:1937-50 (2010) [PubMed] Article Oger, F; Lecorgne, A; Sala, E; Nardese, V; Demay, F; Chevance, S; Desravines, DC; Aleksandrova, N; Le Guével, R; Lorenzi, S; Beccari, AR; Barath, P; Hart, DJ; Bondon, A; Carettoni, D; Simonneaux, G; Salbert, G Biological and biophysical properties of the histone deacetylase inhibitor suberoylanilide hydroxamic acid are affected by the presence of short alkyl groups on the phenyl ring. J Med Chem53:1937-50 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 3 |
|---|
| Name: | Histone deacetylase 3 |
|---|
| Synonyms: | HD3 | HDAC3 | HDAC3_HUMAN | Histone deacetylase 3 (HDAC3) | Human HDAC3 | RPD3-2 | SMAP45 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 48829.55 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O15379 |
|---|
| Residue: | 428 |
|---|
| Sequence: | MAKTVAYFYDPDVGNFHYGAGHPMKPHRLALTHSLVLHYGLYKKMIVFKPYQASQHDMCR
FHSEDYIDFLQRVSPTNMQGFTKSLNAFNVGDDCPVFPGLFEFCSRYTGASLQGATQLNN
KICDIAINWAGGLHHAKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDIHHGDGVQEA
FYLTDRVMTVSFHKYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYKHLFQPVI
NQVVDFYQPTCIVLQCGADSLGCDRLGCFNLSIRGHGECVEYVKSFNIPLLVLGGGGYTV
RNVARCWTYETSLLVEEAISEELPYSEYFEYFAPDFTLHPDVSTRIENQNSRQYLDQIRQ
TIFENLKMLNHAPSVQIHDVPADLLTYDRTDEADAEERGPEENYSRPEAPNEFYDGDHDN
DKESDVEI
|
|
|
|---|
| BDBM50314136 |
|---|
| n/a |
|---|
| Name | BDBM50314136 |
|---|
| Synonyms: | CHEMBL1089339 | N-Hydroxy-N'-(3-methylphenyl)octanediamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H18N2O3 |
|---|
| Mol. Mass. | 250.2936 |
|---|
| SMILES | Cc1cccc(NC(=O)CCCCC(=O)NO)c1 |
|---|
| Structure | 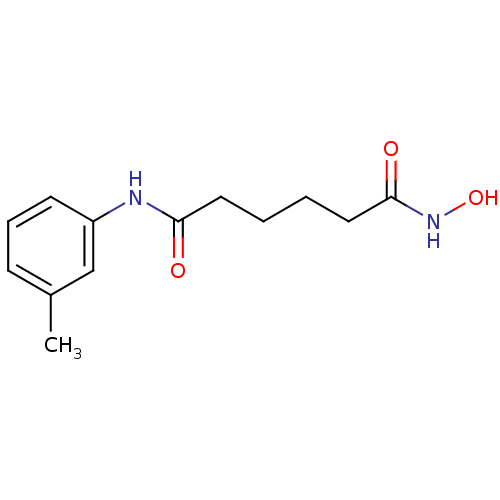
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Oger, F; Lecorgne, A; Sala, E; Nardese, V; Demay, F; Chevance, S; Desravines, DC; Aleksandrova, N; Le Guével, R; Lorenzi, S; Beccari, AR; Barath, P; Hart, DJ; Bondon, A; Carettoni, D; Simonneaux, G; Salbert, G Biological and biophysical properties of the histone deacetylase inhibitor suberoylanilide hydroxamic acid are affected by the presence of short alkyl groups on the phenyl ring. J Med Chem53:1937-50 (2010) [PubMed] Article
Oger, F; Lecorgne, A; Sala, E; Nardese, V; Demay, F; Chevance, S; Desravines, DC; Aleksandrova, N; Le Guével, R; Lorenzi, S; Beccari, AR; Barath, P; Hart, DJ; Bondon, A; Carettoni, D; Simonneaux, G; Salbert, G Biological and biophysical properties of the histone deacetylase inhibitor suberoylanilide hydroxamic acid are affected by the presence of short alkyl groups on the phenyl ring. J Med Chem53:1937-50 (2010) [PubMed] Article