

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | ||
| Ligand | BDBM50323728 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_646800 (CHEMBL1216941) | ||
| IC50 | 220±n/a nM | ||
| Citation |  Berndt, A; Miller, S; Williams, O; Le, DD; Houseman, BT; Pacold, JI; Gorrec, F; Hon, WC; Liu, Y; Rommel, C; Gaillard, P; Rückle, T; Schwarz, MK; Shokat, KM; Shaw, JP; Williams, RL The p110 delta structure: mechanisms for selectivity and potency of new PI(3)K inhibitors. Nat Chem Biol6:117-24 (2010) [PubMed] Article Berndt, A; Miller, S; Williams, O; Le, DD; Houseman, BT; Pacold, JI; Gorrec, F; Hon, WC; Liu, Y; Rommel, C; Gaillard, P; Rückle, T; Schwarz, MK; Shokat, KM; Shaw, JP; Williams, RL The p110 delta structure: mechanisms for selectivity and potency of new PI(3)K inhibitors. Nat Chem Biol6:117-24 (2010) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | |||
| Name: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | ||
| Synonyms: | PI3-kinase p110 subunit beta | PI3-kinase subunit p110-beta | PI3Kbeta | PIK3C1 | PIK3CB | PK3CB_HUMAN | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta (PI3Kbeta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform (PI3K beta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform (PI3K) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform (PI3K-beta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform (PI3Kbeta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform (PI3Kÿ²) | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform | Phosphoinositide 3-Kinase (PI3K), beta | Phosphoinositide 3-Kinase (PI3K), beta Chain A | Phosphoinositide-3-kinase (PI3K beta) | PtdIns-3-kinase p110 | ||
| Type: | Enzyme Subunit | ||
| Mol. Mass.: | 122769.00 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P42338 | ||
| Residue: | 1070 | ||
| Sequence: |
| ||
| BDBM50323728 | |||
| n/a | |||
| Name | BDBM50323728 | ||
| Synonyms: | 2-((4-amino-3-(3-fluoro-5-hydroxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)methyl)-5-methyl-3-o-tolylquinazolin-4(3H)-one | 2-{[4-amino-3-(3-fluoro-5-hydroxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]methyl}-5-methyl-3-(2-methylphenyl)quinazolin-4(3H)-one | CHEMBL1213084 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C28H22FN7O2 | ||
| Mol. Mass. | 507.5184 | ||
| SMILES | Cc1ccccc1-n1c(Cn2nc(-c3cc(O)cc(F)c3)c3c(N)ncnc23)nc2cccc(C)c2c1=O |(2.13,-14.38,;2.12,-12.84,;3.46,-12.06,;3.45,-10.52,;2.1,-9.75,;.77,-10.55,;.79,-12.08,;-.54,-12.85,;-.54,-14.41,;.81,-15.18,;.81,-16.72,;-.43,-17.63,;.05,-19.1,;-.74,-20.41,;-2.27,-20.38,;-3.06,-21.69,;-4.6,-21.66,;-2.32,-23.04,;-.78,-23.06,;-.03,-24.4,;.01,-21.74,;1.59,-19.09,;2.62,-20.22,;2.15,-21.69,;4.12,-19.9,;4.59,-18.43,;3.56,-17.3,;2.07,-17.63,;-1.87,-15.18,;-3.21,-14.41,;-4.55,-15.19,;-5.88,-14.42,;-5.87,-12.87,;-4.55,-12.1,;-4.56,-10.56,;-3.21,-12.87,;-1.88,-12.1,;-1.89,-10.56,)| | ||
| Structure | 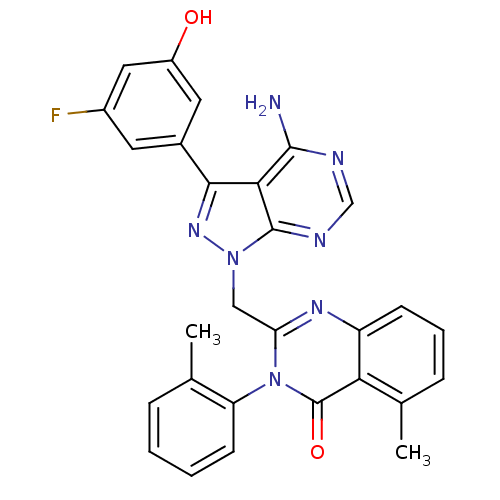
| ||