| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Tyrosine-protein phosphatase non-receptor type 22 |
|---|
| Ligand | BDBM50334193 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_698317 (CHEMBL1646913) |
|---|
| Ki | 7000±n/a nM |
|---|
| Citation |  Karver, CE; Ahmed, VF; Barrios, AM Oxidative inactivation of the lymphoid tyrosine phosphatase mediated by both general and active site directed NO donors. Bioorg Med Chem Lett21:285-7 (2010) [PubMed] Article Karver, CE; Ahmed, VF; Barrios, AM Oxidative inactivation of the lymphoid tyrosine phosphatase mediated by both general and active site directed NO donors. Bioorg Med Chem Lett21:285-7 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Tyrosine-protein phosphatase non-receptor type 22 |
|---|
| Name: | Tyrosine-protein phosphatase non-receptor type 22 |
|---|
| Synonyms: | 3.1.3.48 | Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP | LyP | Lymphoid phosphatase | Lymphoid phosphatase (Lyp) | PEP | PEST-domain phosphatase | PTN22_HUMAN | PTPN22 | PTPN8 | Tyrosine-protein phosphatase non-receptor type 22 (LYP) | tyrosine-protein phosphatase non-receptor type 22 isoform 1 |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | 91712.31 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9Y2R2 |
|---|
| Residue: | 807 |
|---|
| Sequence: | MDQREILQKFLDEAQSKKITKEEFANEFLKLKRQSTKYKADKTYPTTVAEKPKNIKKNRY
KDILPYDYSRVELSLITSDEDSSYINANFIKGVYGPKAYIATQGPLSTTLLDFWRMIWEY
SVLIIVMACMEYEMGKKKCERYWAEPGEMQLEFGPFSVSCEAEKRKSDYIIRTLKVKFNS
ETRTIYQFHYKNWPDHDVPSSIDPILELIWDVRCYQEDDSVPICIHCSAGCGRTGVICAI
DYTWMLLKDGIIPENFSVFSLIREMRTQRPSLVQTQEQYELVYNAVLELFKRQMDVIRDK
HSGTESQAKHCIPEKNHTLQADSYSPNLPKSTTKAAKMMNQQRTKMEIKESSSFDFRTSE
ISAKEELVLHPAKSSTSFDFLELNYSFDKNADTTMKWQTKAFPIVGEPLQKHQSLDLGSL
LFEGCSNSKPVNAAGRYFNSKVPITRTKSTPFELIQQRETKEVDSKENFSYLESQPHDSC
FVEMQAQKVMHVSSAELNYSLPYDSKHQIRNASNVKHHDSSALGVYSYIPLVENPYFSSW
PPSGTSSKMSLDLPEKQDGTVFPSSLLPTSSTSLFSYYNSHDSLSLNSPTNISSLLNQES
AVLATAPRIDDEIPPPLPVRTPESFIVVEEAGEFSPNVPKSLSSAVKVKIGTSLEWGGTS
EPKKFDDSVILRPSKSVKLRSPKSELHQDRSSPPPPLPERTLESFFLADEDCMQAQSIET
YSTSYPDTMENSTSSKQTLKTPGKSFTRSKSLKILRNMKKSICNSCPPNKPAESVQSNNS
SSFLNFGFANRFSKPKGPRNPPPTWNI
|
|
|
|---|
| BDBM50334193 |
|---|
| n/a |
|---|
| Name | BDBM50334193 |
|---|
| Synonyms: | CHEMBL1645419 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C65H109N23O26S |
|---|
| Mol. Mass. | 1660.764 |
|---|
| SMILES | [#6]-[#6]-[#6@H](-[#6])-[#6@H](-[#7]-[#6](=O)-[#6@H](-[#6]-[#6](-[#6])-[#6])-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#7]-[#6](=O)-[#6@H](-[#6])-[#7]-[#6](-[#6])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#8])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6](-[#8])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6](-[#7])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#8])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#16]-[#7]=O)-[#6](=O)-[#7]-[#6@@H](-[#6@@H](-[#6])-[#8])-[#6](=O)-[#7]-[#6@@H](-[#6])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#8])=O)-[#6](=O)-[#7]-[#6]-[#6](-[#7])=O |r| |
|---|
| Structure | 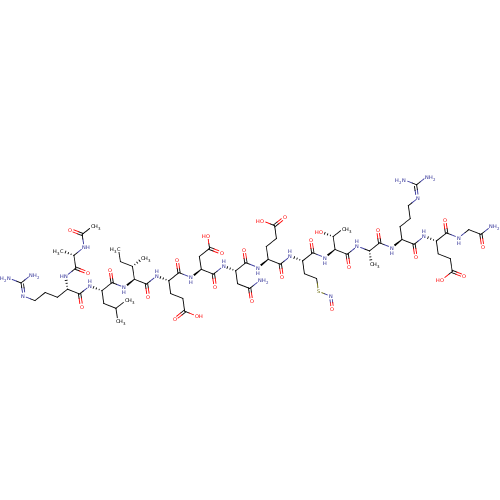
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Karver, CE; Ahmed, VF; Barrios, AM Oxidative inactivation of the lymphoid tyrosine phosphatase mediated by both general and active site directed NO donors. Bioorg Med Chem Lett21:285-7 (2010) [PubMed] Article
Karver, CE; Ahmed, VF; Barrios, AM Oxidative inactivation of the lymphoid tyrosine phosphatase mediated by both general and active site directed NO donors. Bioorg Med Chem Lett21:285-7 (2010) [PubMed] Article