| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Dual specificity calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B |
|---|
| Ligand | BDBM50334670 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_699036 (CHEMBL1646317) |
|---|
| IC50 | 10000±n/a nM |
|---|
| Citation |  Asproni, B; Murineddu, G; Pau, A; Pinna, GA; Langgård, M; Christoffersen, CT; Nielsen, J; Kehler, J Synthesis and SAR study of new phenylimidazole-pyrazolo[1,5-c]quinazolines as potent phosphodiesterase 10A inhibitors. Bioorg Med Chem19:642-9 (2011) [PubMed] Article Asproni, B; Murineddu, G; Pau, A; Pinna, GA; Langgård, M; Christoffersen, CT; Nielsen, J; Kehler, J Synthesis and SAR study of new phenylimidazole-pyrazolo[1,5-c]quinazolines as potent phosphodiesterase 10A inhibitors. Bioorg Med Chem19:642-9 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Dual specificity calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B |
|---|
| Name: | Dual specificity calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B |
|---|
| Synonyms: | 63 kDa Cam-PDE | Calcium/calmodulin-dependent 3 ,5 -cyclic nucleotide phosphodiesterase 1B | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B | Cam-PDE 1B | PDE1B | PDE1B1 | PDE1B_HUMAN | PDES1B | Phosphodiesterase 1B | Phosphodiesterase 1B (PDE1B1) | Phosphodiesterase Type 1 (PDE1B) | Phosphodiesterase, PDE1/PDE5 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 61366.51 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 536 |
|---|
| Sequence: | MELSPRSPPEMLEESDCPSPLELKSAPSKKMWIKLRSLLRYMVKQLENGEINIEELKKNL
EYTASLLEAVYIDETRQILDTEDELQELRSDAVPSEVRDWLASTFTQQARAKGRRAEEKP
KFRSIVHAVQAGIFVERMFRRTYTSVGPTYSTAVLNCLKNLDLWCFDVFSLNQAADDHAL
RTIVFELLTRHNLISRFKIPTVFLMSFLDALETGYGKYKNPYHNQIHAADVTQTVHCFLL
RTGMVHCLSEIELLAIIFAAAIHDYEHTGTTNSFHIQTKSECAIVYNDRSVLENHHISSV
FRLMQDDEMNIFINLTKDEFVELRALVIEMVLATDMSCHFQQVKTMKTALQQLERIDKPK
ALSLLLHAADISHPTKQWLVHSRWTKALMEEFFRQGDKEAELGLPFSPLCDRTSTLVAQS
QIGFIDFIVEPTFSVLTDVAEKSVQPLADEDSKSKNQPSFQWRQPSLDVEVGDPNPDVVS
FRSTWVKRIQENKQKWKERAASGITNQMSIDELSPCEEEAPPSPAEDEHNQNGNLD
|
|
|
|---|
| BDBM50334670 |
|---|
| n/a |
|---|
| Name | BDBM50334670 |
|---|
| Synonyms: | 2,9-Dimethyl-5-(2-(1-methyl-4-phenyl-1H-imidazol-2-yl)ethyl)pyrazolo[1,5-c]quinazoline | CHEMBL1642568 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H23N5 |
|---|
| Mol. Mass. | 381.4729 |
|---|
| SMILES | Cc1cc2c3cc(C)ccc3nc(CCc3nc(cn3C)-c3ccccc3)n2n1 |
|---|
| Structure | 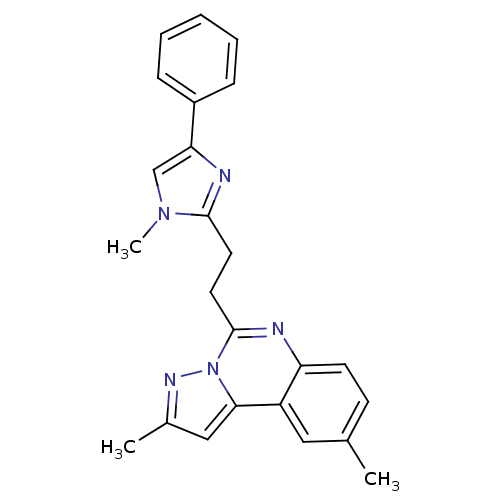
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Asproni, B; Murineddu, G; Pau, A; Pinna, GA; Langgård, M; Christoffersen, CT; Nielsen, J; Kehler, J Synthesis and SAR study of new phenylimidazole-pyrazolo[1,5-c]quinazolines as potent phosphodiesterase 10A inhibitors. Bioorg Med Chem19:642-9 (2011) [PubMed] Article
Asproni, B; Murineddu, G; Pau, A; Pinna, GA; Langgård, M; Christoffersen, CT; Nielsen, J; Kehler, J Synthesis and SAR study of new phenylimidazole-pyrazolo[1,5-c]quinazolines as potent phosphodiesterase 10A inhibitors. Bioorg Med Chem19:642-9 (2011) [PubMed] Article