| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine/threonine-protein kinase PLK1 |
|---|
| Ligand | BDBM50339795 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_735641 (CHEMBL1693343) |
|---|
| IC50 | <9±n/a nM |
|---|
| Citation |  Solanki, S; Innocenti, P; Mas-Droux, C; Boxall, K; Barillari, C; van Montfort, RL; Aherne, GW; Bayliss, R; Hoelder, S Benzimidazole inhibitors induce a DFG-out conformation of never in mitosis gene A-related kinase 2 (Nek2) without binding to the back pocket and reveal a nonlinear structure-activity relationship. J Med Chem54:1626-39 (2011) [PubMed] Article Solanki, S; Innocenti, P; Mas-Droux, C; Boxall, K; Barillari, C; van Montfort, RL; Aherne, GW; Bayliss, R; Hoelder, S Benzimidazole inhibitors induce a DFG-out conformation of never in mitosis gene A-related kinase 2 (Nek2) without binding to the back pocket and reveal a nonlinear structure-activity relationship. J Med Chem54:1626-39 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Serine/threonine-protein kinase PLK1 |
|---|
| Name: | Serine/threonine-protein kinase PLK1 |
|---|
| Synonyms: | PLK | PLK-1 | PLK1 | PLK1_HUMAN | Polo-like kinase 1 (PlK1) | Polo-like kinase 1 (Plk-1) | STPK13 | Serine/threonine-protein kinase (PLK1) | Serine/threonine-protein kinase 13 | polo-like kinase |
|---|
| Type: | Serine/threonine-protein kinase |
|---|
| Mol. Mass.: | 68277.16 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P53350 |
|---|
| Residue: | 603 |
|---|
| Sequence: | MSAAVTAGKLARAPADPGKAGVPGVAAPGAPAAAPPAKEIPEVLVDPRSRRRYVRGRFLG
KGGFAKCFEISDADTKEVFAGKIVPKSLLLKPHQREKMSMEISIHRSLAHQHVVGFHGFF
EDNDFVFVVLELCRRRSLLELHKRRKALTEPEARYYLRQIVLGCQYLHRNRVIHRDLKLG
NLFLNEDLEVKIGDFGLATKVEYDGERKKTLCGTPNYIAPEVLSKKGHSFEVDVWSIGCI
MYTLLVGKPPFETSCLKETYLRIKKNEYSIPKHINPVAASLIQKMLQTDPTARPTINELL
NDEFFTSGYIPARLPITCLTIPPRFSIAPSSLDPSNRKPLTVLNKGLENPLPERPREKEE
PVVRETGEVVDCHLSDMLQQLHSVNASKPSERGLVRQEEAEDPACIPIFWVSKWVDYSDK
YGLGYQLCDNSVGVLFNDSTRLILYNDGDSLQYIERDGTESYLTVSSHPNSLMKKITLLK
YFRNYMSEHLLKAGANITPREGDELARLPYLRTWFRTRSAIILHLSNGSVQINFFQDHTK
LILCPLMAAVTYIDEKRDFRTYRLSLLEEYGCCKELASRLRYARTMVDKLLSSRSASNRL
KAS
|
|
|
|---|
| BDBM50339795 |
|---|
| n/a |
|---|
| Name | BDBM50339795 |
|---|
| Synonyms: | 5-(6-(1-Methylpiperidin-4-yloxy)-1H-benzo[d]imidazol-1-yl)-3-(2-(trifluoromethyl)benzyloxy)thiophene-2-carboxamide | 5-{6-[(1-methylpiperidin-4-yl)oxy]-1H-benzimidazol-1-yl}-3-{[2-(trifluoromethyl)benzyl]oxy}thiophene-2-carboxamide | CHEMBL1614933 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H25F3N4O3S |
|---|
| Mol. Mass. | 530.562 |
|---|
| SMILES | CN1CCC(CC1)Oc1ccc2ncn(-c3cc(OCc4ccccc4C(F)(F)F)c(s3)C(N)=O)c2c1 |
|---|
| Structure | 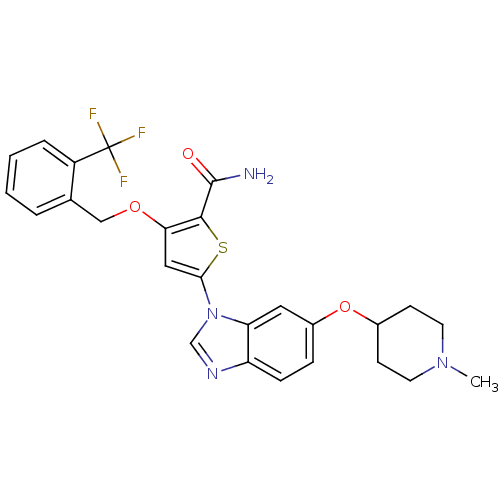
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Solanki, S; Innocenti, P; Mas-Droux, C; Boxall, K; Barillari, C; van Montfort, RL; Aherne, GW; Bayliss, R; Hoelder, S Benzimidazole inhibitors induce a DFG-out conformation of never in mitosis gene A-related kinase 2 (Nek2) without binding to the back pocket and reveal a nonlinear structure-activity relationship. J Med Chem54:1626-39 (2011) [PubMed] Article
Solanki, S; Innocenti, P; Mas-Droux, C; Boxall, K; Barillari, C; van Montfort, RL; Aherne, GW; Bayliss, R; Hoelder, S Benzimidazole inhibitors induce a DFG-out conformation of never in mitosis gene A-related kinase 2 (Nek2) without binding to the back pocket and reveal a nonlinear structure-activity relationship. J Med Chem54:1626-39 (2011) [PubMed] Article