| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cannabinoid receptor 1 |
|---|
| Ligand | BDBM50335937 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_761450 (CHEMBL1817171) |
|---|
| Ki | >10000±n/a nM |
|---|
| Citation |  Pasquini, S; De Rosa, M; Pedani, V; Mugnaini, C; Guida, F; Luongo, L; De Chiaro, M; Maione, S; Dragoni, S; Frosini, M; Ligresti, A; Di Marzo, V; Corelli, F Investigations on the 4-quinolone-3-carboxylic acid motif. 4. Identification of new potent and selective ligands for the cannabinoid type 2 receptor with diverse substitution patterns and antihyperalgesic effects in mice. J Med Chem54:5444-53 (2011) [PubMed] Article Pasquini, S; De Rosa, M; Pedani, V; Mugnaini, C; Guida, F; Luongo, L; De Chiaro, M; Maione, S; Dragoni, S; Frosini, M; Ligresti, A; Di Marzo, V; Corelli, F Investigations on the 4-quinolone-3-carboxylic acid motif. 4. Identification of new potent and selective ligands for the cannabinoid type 2 receptor with diverse substitution patterns and antihyperalgesic effects in mice. J Med Chem54:5444-53 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cannabinoid receptor 1 |
|---|
| Name: | Cannabinoid receptor 1 |
|---|
| Synonyms: | CANN6 | CANNABINOID CB1 | CB-R | CB1 | CNR | CNR1 | CNR1_HUMAN | Cannabinoid CB1 receptor | Cannabinoid receptor | Cannabinoid receptor 1 (CB1) | Cannabinoid receptor 1 (brain) |
|---|
| Type: | G Protein-Coupled Receptor (GPCR) |
|---|
| Mol. Mass.: | 52868.96 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P21554 |
|---|
| Residue: | 472 |
|---|
| Sequence: | MKSILDGLADTTFRTITTDLLYVGSNDIQYEDIKGDMASKLGYFPQKFPLTSFRGSPFQE
KMTAGDNPQLVPADQVNITEFYNKSLSSFKENEENIQCGENFMDIECFMVLNPSQQLAIA
VLSLTLGTFTVLENLLVLCVILHSRSLRCRPSYHFIGSLAVADLLGSVIFVYSFIDFHVF
HRKDSRNVFLFKLGGVTASFTASVGSLFLTAIDRYISIHRPLAYKRIVTRPKAVVAFCLM
WTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWK
AHSHAVRMIQRGTQKSIIIHTSEDGKVQVTRPDQARMDIRLAKTLVLILVVLIICWGPLL
AIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMFPSCEGTAQ
PLDNSMGDSDCLHKHANNAASVHRAAESCIKSTVKIAKVTMSVSTDTSAEAL
|
|
|
|---|
| BDBM50335937 |
|---|
| n/a |
|---|
| Name | BDBM50335937 |
|---|
| Synonyms: | 8-Methyl-4-oxo-1-pentyl-1,4-dihydro-quinoline-3-carboxylic acid adamantan-1-ylamide | CHEMBL1668518 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H34N2O2 |
|---|
| Mol. Mass. | 406.5604 |
|---|
| SMILES | CCCCCn1cc(C(=O)NC23CC4CC(CC(C4)C2)C3)c(=O)c2cccc(C)c12 |TLB:10:11:14:18.16.17,THB:16:15:12:18.17.19,16:17:14.15.20:12,19:17:14:20.11.12,19:11:14:18.16.17| |
|---|
| Structure | 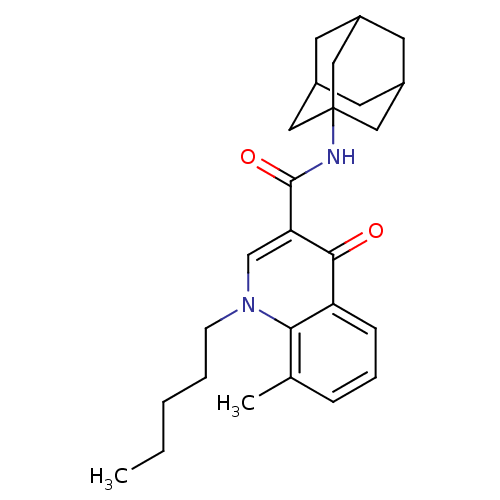
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Pasquini, S; De Rosa, M; Pedani, V; Mugnaini, C; Guida, F; Luongo, L; De Chiaro, M; Maione, S; Dragoni, S; Frosini, M; Ligresti, A; Di Marzo, V; Corelli, F Investigations on the 4-quinolone-3-carboxylic acid motif. 4. Identification of new potent and selective ligands for the cannabinoid type 2 receptor with diverse substitution patterns and antihyperalgesic effects in mice. J Med Chem54:5444-53 (2011) [PubMed] Article
Pasquini, S; De Rosa, M; Pedani, V; Mugnaini, C; Guida, F; Luongo, L; De Chiaro, M; Maione, S; Dragoni, S; Frosini, M; Ligresti, A; Di Marzo, V; Corelli, F Investigations on the 4-quinolone-3-carboxylic acid motif. 4. Identification of new potent and selective ligands for the cannabinoid type 2 receptor with diverse substitution patterns and antihyperalgesic effects in mice. J Med Chem54:5444-53 (2011) [PubMed] Article