

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Histamine H3 receptor | ||
| Ligand | BDBM50352087 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_766550 (CHEMBL1827031) | ||
| Ki | 0.41±n/a nM | ||
| Citation |  Labeeuw, O; Levoin, N; Poupardin-Olivier, O; Calmels, T; Ligneau, X; Berrebi-Bertrand, I; Robert, P; Lecomte, JM; Schwartz, JC; Capet, M Novel and highly potent histamine H3 receptor ligands. Part 2: exploring the cyclohexylamine-based series. Bioorg Med Chem Lett21:5384-8 (2011) [PubMed] Article Labeeuw, O; Levoin, N; Poupardin-Olivier, O; Calmels, T; Ligneau, X; Berrebi-Bertrand, I; Robert, P; Lecomte, JM; Schwartz, JC; Capet, M Novel and highly potent histamine H3 receptor ligands. Part 2: exploring the cyclohexylamine-based series. Bioorg Med Chem Lett21:5384-8 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Histamine H3 receptor | |||
| Name: | Histamine H3 receptor | ||
| Synonyms: | G-protein coupled receptor 97 | GPCR97 | HH3R | HISTAMINE H3 | HRH3 | HRH3_HUMAN | Histamine H3 receptor (H3) | Histamine H3L | Histamine receptor (H3 and H4) | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 48691.47 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Binding assays were using CHO cells stably expressing hH3R receptors. | ||
| Residue: | 445 | ||
| Sequence: |
| ||
| BDBM50352087 | |||
| n/a | |||
| Name | BDBM50352087 | ||
| Synonyms: | CHEMBL1824244 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C28H38N2O | ||
| Mol. Mass. | 418.6141 | ||
| SMILES | C(COc1ccc(cc1)[C@@H]1CC[C@@H](CC1)N1Cc2ccccc2C1)CN1CCCCC1 |r,wU:9.9,12.16,(-4.94,-32.61,;-3.4,-32.61,;-2.63,-31.28,;-1.09,-31.28,;-.33,-32.62,;1.2,-32.63,;1.98,-31.3,;1.22,-29.96,;-.33,-29.95,;3.53,-31.31,;4.29,-32.65,;5.82,-32.67,;6.61,-31.34,;5.84,-30,;4.3,-29.98,;8.15,-31.35,;9.04,-32.61,;10.5,-32.14,;11.83,-32.92,;13.17,-32.17,;13.18,-30.62,;11.85,-29.85,;10.52,-30.6,;9.06,-30.11,;-5.71,-31.27,;-7.25,-31.27,;-8.02,-32.6,;-9.56,-32.6,;-10.33,-31.27,;-9.56,-29.94,;-8.02,-29.94,)| | ||
| Structure | 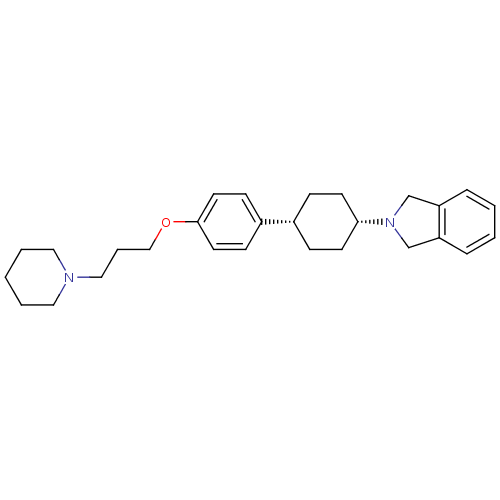
| ||