| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mitogen-activated protein kinase 8 |
|---|
| Ligand | BDBM50352630 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_767683 (CHEMBL1825795) |
|---|
| IC50 | 10000±n/a nM |
|---|
| Citation |  Stebbins, JL; De, SK; Pavlickova, P; Chen, V; Machleidt, T; Chen, LH; Kuntzen, C; Kitada, S; Karin, M; Pellecchia, M Design and characterization of a potent and selective dual ATP- and substrate-competitive subnanomolar bidentate c-Jun N-terminal kinase (JNK) inhibitor. J Med Chem54:6206-14 (2011) [PubMed] Article Stebbins, JL; De, SK; Pavlickova, P; Chen, V; Machleidt, T; Chen, LH; Kuntzen, C; Kitada, S; Karin, M; Pellecchia, M Design and characterization of a potent and selective dual ATP- and substrate-competitive subnanomolar bidentate c-Jun N-terminal kinase (JNK) inhibitor. J Med Chem54:6206-14 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Mitogen-activated protein kinase 8 |
|---|
| Name: | Mitogen-activated protein kinase 8 |
|---|
| Synonyms: | JNK-46 | JNK1 | JNK1-alpha-1 | MAPK8 | MK08_HUMAN | Mitogen-Activated Protein Kinase 8 (JNK1) | PRKM8 | SAPK1 | SAPK1C | Stress-activated protein kinase JNK1 | c-Jun N-terminal kinase 1 | c-Jun N-terminal kinase 1 (JNK1) | c-Jun N-terminal kinase 1(JNK1) | c-Jun N-terminal kinase 2 (JNK2) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 48297.57 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | JNK-1 was purchased from Upstate Cell Signaling Solutions (formerly Upstate Biotechnology). |
|---|
| Residue: | 427 |
|---|
| Sequence: | MSRSKRDNNFYSVEIGDSTFTVLKRYQNLKPIGSGAQGIVCAAYDAILERNVAIKKLSRP
FQNQTHAKRAYRELVLMKCVNHKNIIGLLNVFTPQKSLEEFQDVYIVMELMDANLCQVIQ
MELDHERMSYLLYQMLCGIKHLHSAGIIHRDLKPSNIVVKSDCTLKILDFGLARTAGTSF
MMTPYVVTRYYRAPEVILGMGYKENVDLWSVGCIMGEMVCHKILFPGRDYIDQWNKVIEQ
LGTPCPEFMKKLQPTVRTYVENRPKYAGYSFEKLFPDVLFPADSEHNKLKASQARDLLSK
MLVIDASKRISVDEALQHPYINVWYDPSEAEAPPPKIPDKQLDEREHTIEEWKELIYKEV
MDLEERTKNGVIRGQPSPLGAAVINGSQHPSSSSSVNDVSSMSTDPTLASDTDSSLEAAA
GPLGCCR
|
|
|
|---|
| BDBM50352630 |
|---|
| n/a |
|---|
| Name | BDBM50352630 |
|---|
| Synonyms: | CHEMBL1822310 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C170H295N69O43 |
|---|
| Mol. Mass. | 3993.5978 |
|---|
| SMILES | [#6]-[#6](-[#6])-[#6]-[#6@@H](-[#7]-[#6](=O)-[#6@@H](-[#6]-c1ccccc1)-[#7]-[#6](=O)-[#6@H]-1-[#6]-[#6]-[#6]-[#7]-1-[#6](=O)-[#6@@H](-[#6]-[#6]-[#6](-[#7])=O)-[#7]-[#6](=O)-[#6@H](-[#7]-[#6](=O)-[#6@H]-1-[#6]-[#6]-[#6]-[#7]-1-[#6](=O)-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#7]-[#6](=O)-[#6@@H](-[#6]-[#8])-[#7]-[#6](=O)-[#6@@H](-[#6]-[#6]-[#6](-[#7])=O)-[#7]-[#6](=O)-[#6@@H](-[#6]-[#6](-[#8])=O)-[#7]-[#6](=O)-[#6@H](-[#7]-[#6](-[#6])=O)-[#6@@H](-[#6])-[#8])-[#6](-[#6])-[#6])-[#6](=O)-[#7]-[#6@H](-[#6]-[#6](-[#7])=O)-[#6](=O)-[#7]-[#6@H](-[#6]-[#6](-[#6])-[#6])-[#6](=O)-[#7]-[#6@H](-[#6@H](-[#6])-[#8])-[#6](=O)-[#7]-[#6@H](-[#6@H](-[#6])-[#8])-[#6](=O)-[#7]-1-[#6]-[#6]-[#6]-[#6@@H]-1-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-1-[#6]-[#6]-[#6]-[#6@@H]-1-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-1-[#6]-[#6]-[#6]-[#6@@H]-1-[#6](=O)-[#7]-1-[#6]-[#6]-[#6]-[#6@@H]-1-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]-[#7]-[#6](-[#7])=[#7])-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]-[#7]-[#6](-[#7])=[#7])-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]-[#7]-[#6](-[#7])=[#7])-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6](-[#7])=O)-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]-[#7]-[#6](-[#7])=[#7])-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]-[#7]-[#6](-[#7])=[#7])-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-[#6]-[#6](-[#7])=O |r| |
|---|
| Structure | 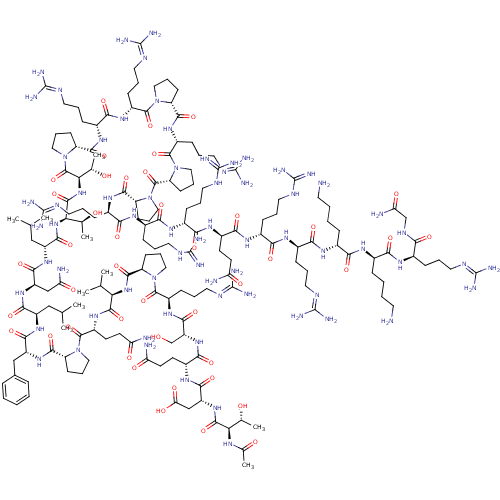
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Stebbins, JL; De, SK; Pavlickova, P; Chen, V; Machleidt, T; Chen, LH; Kuntzen, C; Kitada, S; Karin, M; Pellecchia, M Design and characterization of a potent and selective dual ATP- and substrate-competitive subnanomolar bidentate c-Jun N-terminal kinase (JNK) inhibitor. J Med Chem54:6206-14 (2011) [PubMed] Article
Stebbins, JL; De, SK; Pavlickova, P; Chen, V; Machleidt, T; Chen, LH; Kuntzen, C; Kitada, S; Karin, M; Pellecchia, M Design and characterization of a potent and selective dual ATP- and substrate-competitive subnanomolar bidentate c-Jun N-terminal kinase (JNK) inhibitor. J Med Chem54:6206-14 (2011) [PubMed] Article