| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Matrix metalloproteinase-14 |
|---|
| Ligand | BDBM50148216 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_106790 (CHEMBL873933) |
|---|
| IC50 | 7.4±n/a nM |
|---|
| Citation |  Reiter, LA; Robinson, RP; McClure, KF; Jones, CS; Reese, MR; Mitchell, PG; Otterness, IG; Bliven, ML; Liras, J; Cortina, SR; Donahue, KM; Eskra, JD; Griffiths, RJ; Lame, ME; Lopez-Anaya, A; Martinelli, GJ; McGahee, SM; Yocum, SA; Lopresti-Morrow, LL; Tobiassen, LM; Vaughn-Bowser, ML Pyran-containing sulfonamide hydroxamic acids: potent MMP inhibitors that spare MMP-1. Bioorg Med Chem Lett14:3389-95 (2004) [PubMed] Article Reiter, LA; Robinson, RP; McClure, KF; Jones, CS; Reese, MR; Mitchell, PG; Otterness, IG; Bliven, ML; Liras, J; Cortina, SR; Donahue, KM; Eskra, JD; Griffiths, RJ; Lame, ME; Lopez-Anaya, A; Martinelli, GJ; McGahee, SM; Yocum, SA; Lopresti-Morrow, LL; Tobiassen, LM; Vaughn-Bowser, ML Pyran-containing sulfonamide hydroxamic acids: potent MMP inhibitors that spare MMP-1. Bioorg Med Chem Lett14:3389-95 (2004) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Matrix metalloproteinase-14 |
|---|
| Name: | Matrix metalloproteinase-14 |
|---|
| Synonyms: | MMP-14 | MMP-X1 | MMP14 | MMP14_HUMAN | MT-MMP 1 | MT1-MMP | MT1MMP | MTMMP1 | Matrix Metalloproteinase-14 (MMP-14) | Matrix metalloproteinase 14 | Matrix metalloproteinase-14 | Matrix metalloproteinase-14 (MMP14) | Membrane-type matrix metalloproteinase 1 | Membrane-type-1 matrix metalloproteinase |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 65900.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P50281 |
|---|
| Residue: | 582 |
|---|
| Sequence: | MSPAPRPPRCLLLPLLTLGTALASLGSAQSSSFSPEAWLQQYGYLPPGDLRTHTQRSPQS
LSAAIAAMQKFYGLQVTGKADADTMKAMRRPRCGVPDKFGAEIKANVRRKRYAIQGLKWQ
HNEITFCIQNYTPKVGEYATYEAIRKAFRVWESATPLRFREVPYAYIREGHEKQADIMIF
FAEGFHGDSTPFDGEGGFLAHAYFPGPNIGGDTHFDSAEPWTVRNEDLNGNDIFLVAVHE
LGHALGLEHSSDPSAIMAPFYQWMDTENFVLPDDDRRGIQQLYGGESGFPTKMPPQPRTT
SRPSVPDKPKNPTYGPNICDGNFDTVAMLRGEMFVFKERWFWRVRNNQVMDGYPMPIGQF
WRGLPASINTAYERKDGKFVFFKGDKHWVFDEASLEPGYPKHIKELGRGLPTDKIDAALF
WMPNGKTYFFRGNKYYRFNEELRAVDSEYPKNIKVWEGIPESPRGSFMGSDEVFTYFYKG
NKYWKFNNQKLKVEPGYPKSALRDWMGCPSGGRPDEGTEEETEVIIIEVDEEGGGAVSAA
AVVLPVLLLLLVLAVGLAVFFFRRHGTPRRLLYCQRSLLDKV
|
|
|
|---|
| BDBM50148216 |
|---|
| n/a |
|---|
| Name | BDBM50148216 |
|---|
| Synonyms: | 4-[4-(4-Fluoro-phenoxy)-benzenesulfonylamino]-tetrahydro-pyran-4-carboxylic acid hydroxyamide | CHEMBL111856 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H19FN2O6S |
|---|
| Mol. Mass. | 410.417 |
|---|
| SMILES | ONC(=O)C1(CCOCC1)NS(=O)(=O)c1ccc(Oc2ccc(F)cc2)cc1 |
|---|
| Structure | 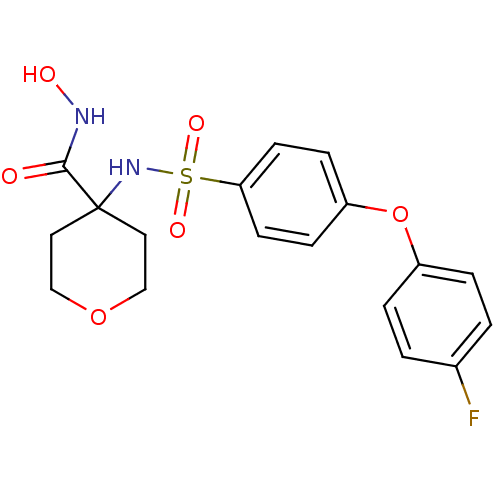
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Reiter, LA; Robinson, RP; McClure, KF; Jones, CS; Reese, MR; Mitchell, PG; Otterness, IG; Bliven, ML; Liras, J; Cortina, SR; Donahue, KM; Eskra, JD; Griffiths, RJ; Lame, ME; Lopez-Anaya, A; Martinelli, GJ; McGahee, SM; Yocum, SA; Lopresti-Morrow, LL; Tobiassen, LM; Vaughn-Bowser, ML Pyran-containing sulfonamide hydroxamic acids: potent MMP inhibitors that spare MMP-1. Bioorg Med Chem Lett14:3389-95 (2004) [PubMed] Article
Reiter, LA; Robinson, RP; McClure, KF; Jones, CS; Reese, MR; Mitchell, PG; Otterness, IG; Bliven, ML; Liras, J; Cortina, SR; Donahue, KM; Eskra, JD; Griffiths, RJ; Lame, ME; Lopez-Anaya, A; Martinelli, GJ; McGahee, SM; Yocum, SA; Lopresti-Morrow, LL; Tobiassen, LM; Vaughn-Bowser, ML Pyran-containing sulfonamide hydroxamic acids: potent MMP inhibitors that spare MMP-1. Bioorg Med Chem Lett14:3389-95 (2004) [PubMed] Article