

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Tyrosine-protein phosphatase non-receptor type 1 | ||
| Ligand | BDBM50143424 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_154471 | ||
| IC50 | 600000±n/a nM | ||
| Citation |  Shrestha, S; Shim, YS; Kim, KC; Lee, KH; Cho, H Evans Blue and other dyes as protein tyrosine phosphatase inhibitors. Bioorg Med Chem Lett14:1923-6 (2004) [PubMed] Article Shrestha, S; Shim, YS; Kim, KC; Lee, KH; Cho, H Evans Blue and other dyes as protein tyrosine phosphatase inhibitors. Bioorg Med Chem Lett14:1923-6 (2004) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Tyrosine-protein phosphatase non-receptor type 1 | |||
| Name: | Tyrosine-protein phosphatase non-receptor type 1 | ||
| Synonyms: | PTN1_HUMAN | PTP1B | PTPN1 | Protein tyrosine phosphatase 1B (PTP1B) | Protein tyrosine phosphatase-1B (PTP1B) | Protein-tyrosine phosphatase 1B | Protein-tyrosine phosphatase 1B (PTP1B) | Tyrosine-protein phosphatase non-receptor type 1 | Tyrosine-protein phosphatase non-receptor type 1 (PTP1B) | ||
| Type: | Protein phosphatase | ||
| Mol. Mass.: | 49963.76 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Human recombinant GST-fusion PTP1B (1-435). | ||
| Residue: | 435 | ||
| Sequence: |
| ||
| BDBM50143424 | |||
| n/a | |||
| Name | BDBM50143424 | ||
| Synonyms: | 2-(3-hydroxy-6-oxo-6H-xanthen-9-yl)benzoic acid | 2-(6-Hydroxy-3-oxo-3H-xanthen-9-yl)-benzoic acid | 2-(6-oxido-3-oxo-3H-xanthen-9-yl)benzoate | 3',6'-dihydroxyspiro[1,3-dihydroisobenzofuran-1,9'-(9'H-xanthene)]-3-one | CHEMBL177756 | FLUORESCEIN | Fluorescite | Funduscein-25 | cid_3383 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H12O5 | ||
| Mol. Mass. | 332.3063 | ||
| SMILES | OC(=O)c1ccccc1-c1c2ccc(O)cc2oc2cc(=O)ccc12 |(-6.39,-28.75,;-5.64,-30.09,;-6.42,-31.42,;-4.1,-30.1,;-4.09,-28.56,;-2.74,-27.79,;-1.41,-28.57,;-1.42,-30.11,;-2.76,-30.87,;-2.77,-32.41,;-1.43,-33.19,;-.1,-32.42,;1.23,-33.2,;1.22,-34.74,;2.54,-35.52,;-.12,-35.5,;-1.44,-34.73,;-2.78,-35.49,;-4.11,-34.72,;-5.43,-35.48,;-6.76,-34.72,;-8.09,-35.49,;-6.76,-33.18,;-5.43,-32.4,;-4.1,-33.18,)| | ||
| Structure | 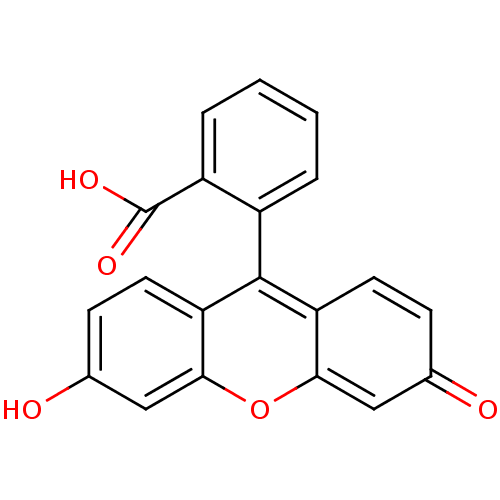
| ||