

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Muscarinic receptor M1 | ||
| Ligand | BDBM50296314 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_140160 | ||
| Ki | 0.200000±n/a nM | ||
| Citation |  Rzeszotarski, WJ; McPherson, DW; Ferkany, JW; Kinnier, WJ; Noronha-Blob, L; Kirkien-Rzeszotarski, A Affinity and selectivity of the optical isomers of 3-quinuclidinyl benzilate and related muscarinic antagonists. J Med Chem31:1463-6 (1988) [PubMed] Rzeszotarski, WJ; McPherson, DW; Ferkany, JW; Kinnier, WJ; Noronha-Blob, L; Kirkien-Rzeszotarski, A Affinity and selectivity of the optical isomers of 3-quinuclidinyl benzilate and related muscarinic antagonists. J Med Chem31:1463-6 (1988) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Muscarinic receptor M1 | |||
| Name: | Muscarinic receptor M1 | ||
| Synonyms: | Muscarinic acetylcholine receptor M1 | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 15022.43 | ||
| Organism: | Bos taurus | ||
| Description: | ChEMBL_140161 | ||
| Residue: | 139 | ||
| Sequence: |
| ||
| BDBM50296314 | |||
| n/a | |||
| Name | BDBM50296314 | ||
| Synonyms: | (3R)-1-Azabicyclo[2.2.2]oct-3-yl9H-xanthene-9-carboxylate | CHEMBL549577 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C21H21NO3 | ||
| Mol. Mass. | 335.3963 | ||
| SMILES | O=C(O[C@H]1CN2CCC1CC2)C1c2ccccc2Oc2ccccc12 |r,wD:3.2,(-1.78,.66,;-3.13,-.09,;-3.15,-1.63,;-1.81,-2.4,;-1.81,-3.94,;-.48,-4.7,;.85,-3.94,;.85,-2.4,;-.48,-1.62,;-.06,-2.86,;-1.11,-3.21,;-4.45,.7,;-5.79,-.04,;-5.82,-1.58,;-7.17,-2.33,;-8.5,-1.53,;-8.47,.02,;-7.11,.76,;-7.09,2.3,;-5.74,3.04,;-5.72,4.57,;-4.38,5.32,;-3.05,4.53,;-3.07,2.99,;-4.42,2.25,)| | ||
| Structure | 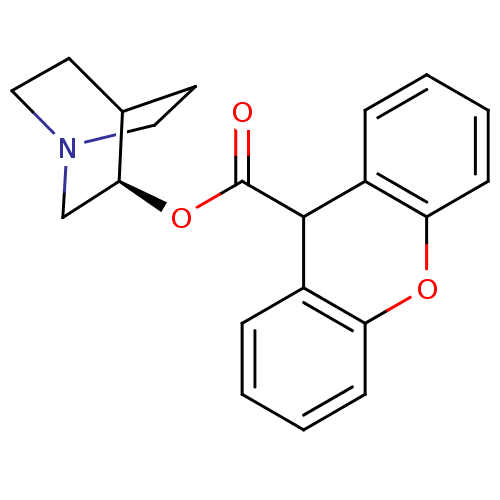
| ||