

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Reverse transcriptase/RNaseH | ||
| Ligand | BDBM50010215 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_195535 (CHEMBL800982) | ||
| IC50 | 43±n/a nM | ||
| Citation |  Silverman, BD; Platt, DE Comparative molecular moment analysis (CoMMA): 3D-QSAR without molecular superposition. J Med Chem39:2129-40 (1996) [PubMed] Article Silverman, BD; Platt, DE Comparative molecular moment analysis (CoMMA): 3D-QSAR without molecular superposition. J Med Chem39:2129-40 (1996) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Reverse transcriptase/RNaseH | |||
| Name: | Reverse transcriptase/RNaseH | ||
| Synonyms: | HIV-1 Reverse Transcriptase RNase H | Human immunodeficiency virus type 1 reverse transcriptase | Reverse transcriptase/RNaseH | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 65229.15 | ||
| Organism: | Human immunodeficiency virus 1 | ||
| Description: | ChEMBL_1473730 | ||
| Residue: | 566 | ||
| Sequence: |
| ||
| BDBM50010215 | |||
| n/a | |||
| Name | BDBM50010215 | ||
| Synonyms: | 4-Chloro-8-methyl-7-(3-methyl-but-2-enyl)-6,7,8,9-tetrahydro-2H-2,7,9a-triaza-benzo[cd]azulene-1-thione | CHEMBL296092 | R(-) 4-Chloro-8-methyl-7-(3-methyl-but-2-enyl)-6,7,8,9-tetrahydro-2H-2,7,9a-triaza-benzo[cd]azulene-1-thione | S(+) 4-Chloro-8-methyl-7-(3-methyl-but-2-enyl)-6,7,8,9-tetrahydro-2H-2,7,9a-triaza-benzo[cd]azulene-1-thione | ||
| Type | Small organic molecule | ||
| Emp. Form. | C16H20ClN3S | ||
| Mol. Mass. | 321.868 | ||
| SMILES | CC1Cn2c3c(CN1CC=C(C)C)cc(Cl)cc3[nH]c2=S |(12.54,-6.89,;11.15,-6.2,;9.93,-7.08,;8.43,-6.69,;7.87,-5.24,;8.57,-3.87,;10.1,-3.61,;11.25,-4.64,;12.66,-4.03,;13.9,-4.98,;15.31,-4.37,;15.5,-2.85,;16.54,-5.31,;7.73,-2.58,;6.2,-2.66,;5.35,-1.38,;5.5,-4.05,;6.34,-5.34,;5.94,-6.82,;7.24,-7.65,;7.33,-9.19,)| | ||
| Structure | 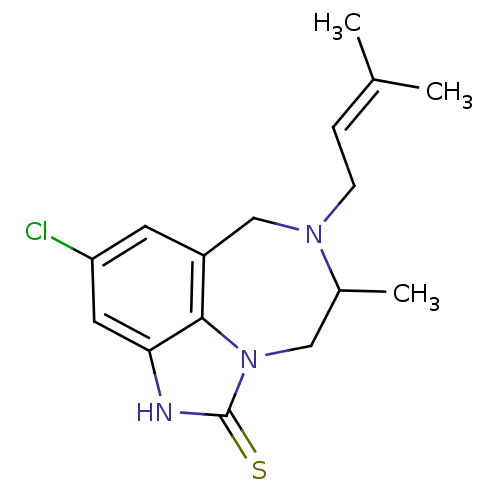
| ||