| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Metabotropic glutamate receptor 6 |
|---|
| Ligand | BDBM17660 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_104311 (CHEMBL718130) |
|---|
| Ki | 1000000±n/a nM |
|---|
| Citation |  Bräuner-Osborne, H; Egebjerg, J; Nielsen, EO; Madsen, U; Krogsgaard-Larsen, P Ligands for glutamate receptors: design and therapeutic prospects. J Med Chem43:2609-45 (2000) [PubMed] Bräuner-Osborne, H; Egebjerg, J; Nielsen, EO; Madsen, U; Krogsgaard-Larsen, P Ligands for glutamate receptors: design and therapeutic prospects. J Med Chem43:2609-45 (2000) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Metabotropic glutamate receptor 6 |
|---|
| Name: | Metabotropic glutamate receptor 6 |
|---|
| Synonyms: | GPRC1F | GRM6 | GRM6_HUMAN | MGLUR6 | Metabotropic glutamate receptor 6 | glutamate receptor, metabotropic 6 precursor | metabotropic glutamate 6 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 95479.26 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | metabotropic glutamate 6 GRM6 HUMAN::O15303 |
|---|
| Residue: | 877 |
|---|
| Sequence: | MARPRRAREPLLVALLPLAWLAQAGLARAAGSVRLAGGLTLGGLFPVHARGAAGRACGQL
KKEQGVHRLEAMLYALDRVNADPELLPGVRLGARLLDTCSRDTYALEQALSFVQALIRGR
GDGDEVGVRCPGGVPPLRPAPPERVVAVVGASASSVSIMVANVLRLFAIPQISYASTAPE
LSDSTRYDFFSRVVPPDSYQAQAMVDIVRALGWNYVSTLASEGNYGESGVEAFVQISREA
GGVCIAQSIKIPREPKPGEFSKVIRRLMETPNARGIIIFANEDDIRRVLEAARQANLTGH
FLWVGSDSWGAKTSPILSLEDVAVGAITILPKRASIDGFDQYFMTRSLENNRRNIWFAEF
WEENFNCKLTSSGTQSDDSTRKCTGEERIGRDSTYEQEGKVQFVIDAVYAIAHALHSMHQ
ALCPGHTGLCPAMEPTDGRMLLQYIRAVRFNGSAGTPVMFNENGDAPGRYDIFQYQATNG
SASSGGYQAVGQWAETLRLDVEALQWSGDPHEVPSSLCSLPCGPGERKKMVKGVPCCWHC
EACDGYRFQVDEFTCEACPGDMRPTPNHTGCRPTPVVRLSWSSPWAAPPLLLAVLGIVAT
TTVVATFVRYNNTPIVRASGRELSYVLLTGIFLIYAITFLMVAEPGAAVCAARRLFLGLG
TTLSYSALLTKTNRIYRIFEQGKRSVTPPPFISPTSQLVITFSLTSLQVVGMIAWLGARP
PHSVIDYEEQRTVDPEQARGVLKCDMSDLSLIGCLGYSLLLMVTCTVYAIKARGVPETFN
EAKPIGFTMYTTCIIWLAFVPIFFGTAQSAEKIYIQTTTLTVSLSLSASVSLGMLYVPKT
YVILFHPEQNVQKRKRSLKATSTVAAPPKGEDAEAHK
|
|
|
|---|
| BDBM17660 |
|---|
| n/a |
|---|
| Name | BDBM17660 |
|---|
| Synonyms: | (2S)-2-amino-3-(3,5-dioxo-1,2,4-oxadiazolidin-2-yl)propanoic acid | 2-amino-3-(3,5-dioxo[1,2,4]oxadiazolidin-2-yl)propionic acid | CHEMBL279956 | Quisqualate | Quisqualate, L | quisqualic acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C5H7N3O5 |
|---|
| Mol. Mass. | 189.1262 |
|---|
| SMILES | N[C@@H](Cn1oc(=O)[nH]c1=O)C(O)=O |
|---|
| Structure | 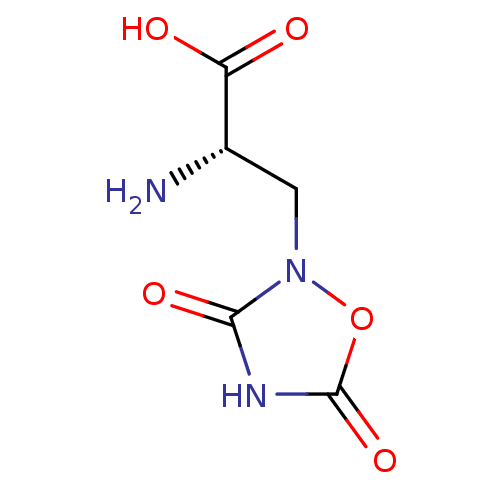
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Bräuner-Osborne, H; Egebjerg, J; Nielsen, EO; Madsen, U; Krogsgaard-Larsen, P Ligands for glutamate receptors: design and therapeutic prospects. J Med Chem43:2609-45 (2000) [PubMed]
Bräuner-Osborne, H; Egebjerg, J; Nielsen, EO; Madsen, U; Krogsgaard-Larsen, P Ligands for glutamate receptors: design and therapeutic prospects. J Med Chem43:2609-45 (2000) [PubMed]