

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | D(2) dopamine receptor | ||
| Ligand | BDBM50007422 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_61937 (CHEMBL675125) | ||
| Kd | 370±n/a nM | ||
| Citation |  Wilcox, RE; Huang, WH; Brusniak, MY; Wilcox, DM; Pearlman, RS; Teeter, MM; DuRand, CJ; Wiens, BL; Neve, KA CoMFA-based prediction of agonist affinities at recombinant wild type versus serine to alanine point mutated D2 dopamine receptors. J Med Chem43:3005-19 (2000) [PubMed] Wilcox, RE; Huang, WH; Brusniak, MY; Wilcox, DM; Pearlman, RS; Teeter, MM; DuRand, CJ; Wiens, BL; Neve, KA CoMFA-based prediction of agonist affinities at recombinant wild type versus serine to alanine point mutated D2 dopamine receptors. J Med Chem43:3005-19 (2000) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| D(2) dopamine receptor | |||
| Name: | D(2) dopamine receptor | ||
| Synonyms: | DOPAMINE D2 | DOPAMINE D2 Long | DOPAMINE D2 Short | DRD2_RAT | Dopamine D2 receptor | Dopamine2-like | Drd2 | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 50931.60 | ||
| Organism: | Rattus norvegicus (rat) | ||
| Description: | P61169 | ||
| Residue: | 444 | ||
| Sequence: |
| ||
| BDBM50007422 | |||
| n/a | |||
| Name | BDBM50007422 | ||
| Synonyms: | (+)-6-Propyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol | (-)-6-Propyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol | (R) 6-Propyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol | (R)-10,11-Dihydroxy-6-propyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinolinium | (R)-6-propyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol | 10,11-Dihydroxy-6-propyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinolinium(R(-)NPA) | 6-Propyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol | CHEMBL225230 | CHEMBL538542 | N-propylapomorphine | N-propylnorapomorphine-(+) | N-propylnorapomorphine-(-) | ||
| Type | Small organic molecule | ||
| Emp. Form. | C19H21NO2 | ||
| Mol. Mass. | 295.3755 | ||
| SMILES | CCCN1CCc2cccc-3c2[C@H]1Cc1ccc(O)c(O)c-31 |r| | ||
| Structure | 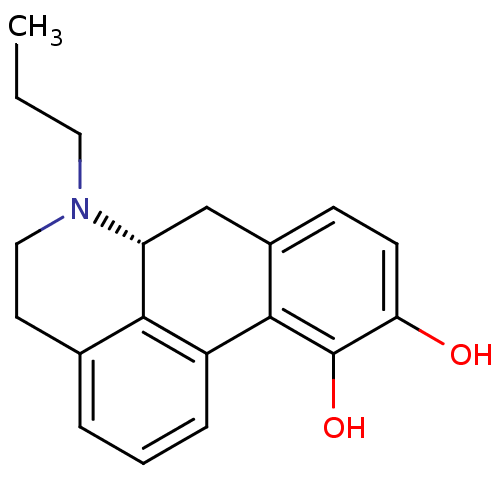
| ||