| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mu-type opioid receptor |
|---|
| Ligand | BDBM50369821 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_145150 (CHEMBL755954) |
|---|
| Ki | 1.5±n/a nM |
|---|
| Citation |  Coop, A; Norton, CL; Berzetei-Gurske, I; Burnside, J; Toll, L; Husbands, SM; Lewis, JW Structural determinants of opioid activity in the orvinols and related structures: ethers of orvinol and isoorvinol. J Med Chem43:1852-7 (2000) [PubMed] Coop, A; Norton, CL; Berzetei-Gurske, I; Burnside, J; Toll, L; Husbands, SM; Lewis, JW Structural determinants of opioid activity in the orvinols and related structures: ethers of orvinol and isoorvinol. J Med Chem43:1852-7 (2000) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Mu-type opioid receptor |
|---|
| Name: | Mu-type opioid receptor |
|---|
| Synonyms: | M-OR-1 | MOR-1 | Mu opioid receptor | Mu-type opioid receptor (Mu) | OPIATE Mu | OPRM1 | OPRM_CAVPO |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 11165.58 |
|---|
| Organism: | GUINEA PIG |
|---|
| Description: | P97266 |
|---|
| Residue: | 98 |
|---|
| Sequence: | YTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGTILCKIVISIDYYNMFTSI
FTLCTMSVDRYIAVCHPVKALDFRTPRNAKTVNVCNWI
|
|
|
|---|
| BDBM50369821 |
|---|
| n/a |
|---|
| Name | BDBM50369821 |
|---|
| Synonyms: | CHEMBL1169585 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H33NO4 |
|---|
| Mol. Mass. | 411.5338 |
|---|
| SMILES | CCC[C@@](C)(O)[C@@H]1CC23C=C[C@]1(OC)[C@H]1Oc4c5c(CC2N(C)CCC315)ccc4O |r,c:9,TLB:16:17:8:21.23.24,3:6:25.14:10.9,24:25:7.6:10.9,22:21:17.18.19:8,THB:17:25:7.6:10.9,7:8:17.18.19:21.23.24,9:8:17.18.19:21.23.24,15:14:7.6:10.9| |
|---|
| Structure | 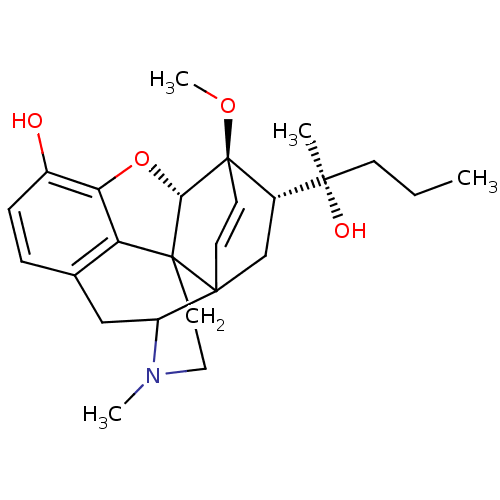
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Coop, A; Norton, CL; Berzetei-Gurske, I; Burnside, J; Toll, L; Husbands, SM; Lewis, JW Structural determinants of opioid activity in the orvinols and related structures: ethers of orvinol and isoorvinol. J Med Chem43:1852-7 (2000) [PubMed]
Coop, A; Norton, CL; Berzetei-Gurske, I; Burnside, J; Toll, L; Husbands, SM; Lewis, JW Structural determinants of opioid activity in the orvinols and related structures: ethers of orvinol and isoorvinol. J Med Chem43:1852-7 (2000) [PubMed]