

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Mu-type opioid receptor | ||
| Ligand | BDBM50100462 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_149630 (CHEMBL756698) | ||
| Ki | 81.8±n/a nM | ||
| Citation |  Sharma, SK; Jones, RM; Metzger, TG; Ferguson, DM; Portoghese, PS Transformation of a kappa-opioid receptor antagonist to a kappa-agonist by transfer of a guanidinium group from the 5'- to 6'-position of naltrindole. J Med Chem44:2073-9 (2001) [PubMed] Sharma, SK; Jones, RM; Metzger, TG; Ferguson, DM; Portoghese, PS Transformation of a kappa-opioid receptor antagonist to a kappa-agonist by transfer of a guanidinium group from the 5'- to 6'-position of naltrindole. J Med Chem44:2073-9 (2001) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Mu-type opioid receptor | |||
| Name: | Mu-type opioid receptor | ||
| Synonyms: | MOR-1 | MUOR1 | Mu-type opioid receptor (MOR) | OPIATE Mu | OPRM_RAT | Opiate non-selective | Opioid receptor B | Oprm1 | Ror-b | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 44503.11 | ||
| Organism: | Rattus norvegicus (rat) | ||
| Description: | Competition binding assays were carried out using membrane preparations from transfected HN9.10 cells that constitutively expressed the mu opioid receptor. | ||
| Residue: | 398 | ||
| Sequence: |
| ||
| BDBM50100462 | |||
| n/a | |||
| Name | BDBM50100462 | ||
| Synonyms: | 6`-guanidino-17-(cyclopropylmethyl)-6,7-didehydro-4,5alpha-epoxy-3,14-hydroxyindolo-[2`,3`:6,7]morphinian | ||
| Type | Small organic molecule | ||
| Emp. Form. | C27H29N5O3 | ||
| Mol. Mass. | 471.5509 | ||
| SMILES | NC(N)=Nc1ccc2c3C[C@@]4(O)C5Cc6ccc(O)c7O[C@@H](c3[nH]c2c1)[C@]4(CCN5CC1CC1)c67 |wU:10.10,wD:26.39,21.20,THB:19:34:10:29.27.28,15:14:10:29.27.28,(16.18,-14.68,;17.06,-15.94,;18.6,-15.8,;16.41,-17.34,;14.87,-17.48,;14,-16.22,;12.47,-16.36,;11.84,-17.76,;10.38,-18.21,;9.06,-17.41,;7.72,-18.16,;8.12,-16.67,;6.39,-17.38,;5.05,-18.14,;5.04,-19.68,;3.72,-20.45,;3.72,-21.99,;5.04,-22.76,;5.04,-24.3,;6.37,-21.99,;8.59,-21.98,;9.01,-20.49,;10.36,-19.75,;11.81,-20.24,;12.72,-19,;14.24,-18.86,;7.68,-19.7,;5.72,-18.42,;5.32,-16.92,;6.42,-15.84,;5.09,-15.06,;5.09,-13.52,;4.32,-12.18,;5.88,-12.19,;6.37,-20.45,)| | ||
| Structure | 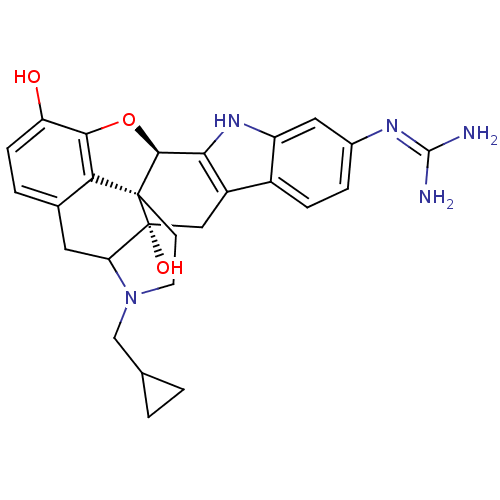
| ||