| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Matrilysin |
|---|
| Ligand | BDBM50102608 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_105053 (CHEMBL711567) |
|---|
| Ki | 1.5±n/a nM |
|---|
| Citation |  Xue, CB; Voss, ME; Nelson, DJ; Duan, JJ; Cherney, RJ; Jacobson, IC; He, X; Roderick, J; Chen, L; Corbett, RL; Wang, L; Meyer, DT; Kennedy, K; DeGradodagger, WF; Hardman, KD; Teleha, CA; Jaffee, BD; Liu, RQ; Copeland, RA; Covington, MB; Christ, DD; Trzaskos, JM; Newton, RC; Magolda, RL; Wexler, RR; Decicco, CP Design, synthesis, and structure-activity relationships of macrocyclic hydroxamic acids that inhibit tumor necrosis factor alpha release in vitro and in vivo. J Med Chem44:2636-60 (2001) [PubMed] Xue, CB; Voss, ME; Nelson, DJ; Duan, JJ; Cherney, RJ; Jacobson, IC; He, X; Roderick, J; Chen, L; Corbett, RL; Wang, L; Meyer, DT; Kennedy, K; DeGradodagger, WF; Hardman, KD; Teleha, CA; Jaffee, BD; Liu, RQ; Copeland, RA; Covington, MB; Christ, DD; Trzaskos, JM; Newton, RC; Magolda, RL; Wexler, RR; Decicco, CP Design, synthesis, and structure-activity relationships of macrocyclic hydroxamic acids that inhibit tumor necrosis factor alpha release in vitro and in vivo. J Med Chem44:2636-60 (2001) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Matrilysin |
|---|
| Name: | Matrilysin |
|---|
| Synonyms: | MMP7 | MMP7_HUMAN | MPSL1 | Matrix metalloproteinase 7 | Matrix metalloproteinase-7 (MMP-7) | Matrix metalloproteinase-7 (MMP7) | PUMP1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 29681.54 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P09237 |
|---|
| Residue: | 267 |
|---|
| Sequence: | MRLTVLCAVCLLPGSLALPLPQEAGGMSELQWEQAQDYLKRFYLYDSETKNANSLEAKLK
EMQKFFGLPITGMLNSRVIEIMQKPRCGVPDVAEYSLFPNSPKWTSKVVTYRIVSYTRDL
PHITVDRLVSKALNMWGKEIPLHFRKVVWGTADIMIGFARGAHGDSYPFDGPGNTLAHAF
APGTGLGGDAHFDEDERWTDGSSLGINFLYAATHELGHSLGMGHSSDPNAVMYPTYGNGD
PQNFKLSQDDIKGIQKLYGKRSNSRKK
|
|
|
|---|
| BDBM50102608 |
|---|
| n/a |
|---|
| Name | BDBM50102608 |
|---|
| Synonyms: | 11-Isobutyl-2,10-dioxo-1-oxa-3,9-diaza-cyclopentadecane-8,12-dicarboxylic acid 12-hydroxyamide 8-[(2-morpholin-4-yl-2-oxo-ethyl)-amide] | 11-Isobutyl-2,10-dioxo-1-oxa-3,9-diaza-cyclopentadecane-8,12-dicarboxylic acid 12-hydroxyamide 8-[(2-morpholin-4-yl-2-oxo-ethyl)-amide](SP057) | CHEMBL91636 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H41N5O8 |
|---|
| Mol. Mass. | 527.611 |
|---|
| SMILES | CC(C)C[C@@H]1[C@H](CCCOC(=O)NCCCC[C@H](NC1=O)C(=O)NCC(=O)N1CCOCC1)C(=O)NO |
|---|
| Structure | 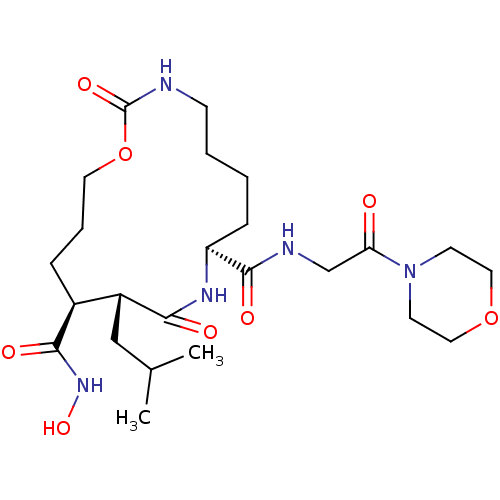
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Xue, CB; Voss, ME; Nelson, DJ; Duan, JJ; Cherney, RJ; Jacobson, IC; He, X; Roderick, J; Chen, L; Corbett, RL; Wang, L; Meyer, DT; Kennedy, K; DeGradodagger, WF; Hardman, KD; Teleha, CA; Jaffee, BD; Liu, RQ; Copeland, RA; Covington, MB; Christ, DD; Trzaskos, JM; Newton, RC; Magolda, RL; Wexler, RR; Decicco, CP Design, synthesis, and structure-activity relationships of macrocyclic hydroxamic acids that inhibit tumor necrosis factor alpha release in vitro and in vivo. J Med Chem44:2636-60 (2001) [PubMed]
Xue, CB; Voss, ME; Nelson, DJ; Duan, JJ; Cherney, RJ; Jacobson, IC; He, X; Roderick, J; Chen, L; Corbett, RL; Wang, L; Meyer, DT; Kennedy, K; DeGradodagger, WF; Hardman, KD; Teleha, CA; Jaffee, BD; Liu, RQ; Copeland, RA; Covington, MB; Christ, DD; Trzaskos, JM; Newton, RC; Magolda, RL; Wexler, RR; Decicco, CP Design, synthesis, and structure-activity relationships of macrocyclic hydroxamic acids that inhibit tumor necrosis factor alpha release in vitro and in vivo. J Med Chem44:2636-60 (2001) [PubMed]