| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fatty-acid amide hydrolase 1 [30-579] |
|---|
| Ligand | BDBM50107607 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_68636 (CHEMBL680041) |
|---|
| IC50 | 30000±n/a nM |
|---|
| Citation |  López-Rodríguez, ML; Viso, A; Ortega-Gutiérrez, S; Fowler, CJ; Tiger, G; de Lago, E; Fernández-Ruiz, J; Ramos, JA Design, synthesis, and biological evaluation of new inhibitors of the endocannabinoid uptake: comparison with effects on fatty acid amidohydrolase. J Med Chem46:1512-22 (2003) [PubMed] Article López-Rodríguez, ML; Viso, A; Ortega-Gutiérrez, S; Fowler, CJ; Tiger, G; de Lago, E; Fernández-Ruiz, J; Ramos, JA Design, synthesis, and biological evaluation of new inhibitors of the endocannabinoid uptake: comparison with effects on fatty acid amidohydrolase. J Med Chem46:1512-22 (2003) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Fatty-acid amide hydrolase 1 [30-579] |
|---|
| Name: | Fatty-acid amide hydrolase 1 [30-579] |
|---|
| Synonyms: | Anandamide amidohydrolase 1 | FAAH1_RAT | Faah | Faah1 | Fatty Acid Amide Hydrolase | Fatty Acid Amide Hydrolic, FAAH | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Fatty-acid amide hydrolase 1 (FAAH) | Fatty-acid amide hydrolase 1 (aa 30-579) | Oleamide hydrolase 1 |
|---|
| Type: | Single-pass membrane protein; homodimer |
|---|
| Mol. Mass.: | 60474.00 |
|---|
| Organism: | Rattus norvegicus (rat) |
|---|
| Description: | P97612 (aa 30-579) |
|---|
| Residue: | 550 |
|---|
| Sequence: | RWTGRQKARGAATRARQKQRASLETMDKAVQRFRLQNPDLDSEALLTLPLLQLVQKLQSG
ELSPEAVFFTYLGKAWEVNKGTNCVTSYLTDCETQLSQAPRQGLLYGVPVSLKECFSYKG
HDSTLGLSLNEGMPSESDCVVVQVLKLQGAVPFVHTNVPQSMLSFDCSNPLFGQTMNPWK
SSKSPGGSSGGEGALIGSGGSPLGLGTDIGGSIRFPSAFCGICGLKPTGNRLSKSGLKGC
VYGQTAVQLSLGPMARDVESLALCLKALLCEHLFTLDPTVPPLPFREEVYRSSRPLRVGY
YETDNYTMPSPAMRRALIETKQRLEAAGHTLIPFLPNNIPYALEVLSAGGLFSDGGRSFL
QNFKGDFVDPCLGDLILILRLPSWFKRLLSLLLKPLFPRLAAFLNSMRPRSAEKLWKLQH
EIEMYRQSVIAQWKAMNLDVLLTPMLGPALDLNTPGRATGAISYTVLYNCLDFPAGVVPV
TTVTAEDDAQMELYKGYFGDIWDIILKKAMKNSVGLPVAVQCVALPWQEELCLRFMREVE
QLMTPQKQPS
|
|
|
|---|
| BDBM50107607 |
|---|
| n/a |
|---|
| Name | BDBM50107607 |
|---|
| Synonyms: | (5Z,8Z)-Icosa-5,8,11,14-tetraenoic acid (furan-3-ylmethyl)-amide | (5Z,8Z,11Z,14Z)-Icosa-5,8,11,14-tetraenoic acid (furan-3-ylmethyl)-amide | CHEMBL434778 | Icosa-5,8,11,14-tetraenoic acid (furan-3-ylmethyl)-amide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H37NO2 |
|---|
| Mol. Mass. | 383.5668 |
|---|
| SMILES | CCCCC\C=C/C\C=C/C\C=C/C\C=C/CCCC(=O)NCc1ccoc1 |
|---|
| Structure | 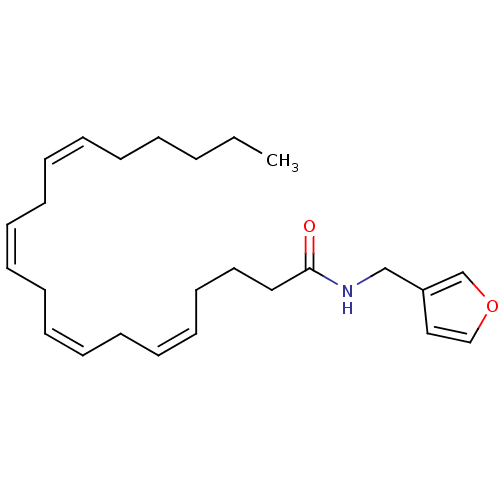
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 López-Rodríguez, ML; Viso, A; Ortega-Gutiérrez, S; Fowler, CJ; Tiger, G; de Lago, E; Fernández-Ruiz, J; Ramos, JA Design, synthesis, and biological evaluation of new inhibitors of the endocannabinoid uptake: comparison with effects on fatty acid amidohydrolase. J Med Chem46:1512-22 (2003) [PubMed] Article
López-Rodríguez, ML; Viso, A; Ortega-Gutiérrez, S; Fowler, CJ; Tiger, G; de Lago, E; Fernández-Ruiz, J; Ramos, JA Design, synthesis, and biological evaluation of new inhibitors of the endocannabinoid uptake: comparison with effects on fatty acid amidohydrolase. J Med Chem46:1512-22 (2003) [PubMed] Article