| Reaction Details |
|---|
 | Report a problem with these data |
| Target | High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A/cAMP-specific 3',5'-cyclic phosphodiesterase 7B |
|---|
| Ligand | BDBM23620 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_302392 (CHEMBL875201) |
|---|
| Ki | 600±n/a nM |
|---|
| Citation |  Manallack, DT; Hughes, RA; Thompson, PE The next generation of phosphodiesterase inhibitors: structural clues to ligand and substrate selectivity of phosphodiesterases. J Med Chem48:3449-62 (2005) [PubMed] Article Manallack, DT; Hughes, RA; Thompson, PE The next generation of phosphodiesterase inhibitors: structural clues to ligand and substrate selectivity of phosphodiesterases. J Med Chem48:3449-62 (2005) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A/cAMP-specific 3',5'-cyclic phosphodiesterase 7B |
|---|
| Name: | High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A/cAMP-specific 3',5'-cyclic phosphodiesterase 7B |
|---|
| Synonyms: | Phosphodiesterase 7 |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of ChEMBL is 302392 |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 7B |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | PDE7B | PDE7B_HUMAN | Phosphodiesterase 7B | Phosphodiesterase 7B (PDE7B) | cAMP-specific 3',5'-cyclic phosphodiesterase 7B |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 51842.76 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9NP56 |
|---|
| Residue: | 450 |
|---|
| Sequence: | MSCLMVERCGEILFENPDQNAKCVCMLGDIRLRGQTGVRAERRGSYPFIDFRLLNSTTYS
GEIGTKKKVKRLLSFQRYFHASRLLRGIIPQAPLHLLDEDYLGQARHMLSKVGMWDFDIF
LFDRLTNGNSLVTLLCHLFNTHGLIHHFKLDMVTLHRFLVMVQEDYHSQNPYHNAVHAAD
VTQAMHCYLKEPKLASFLTPLDIMLGLLAAAAHDVDHPGVNQPFLIKTNHHLANLYQNMS
VLENHHWRSTIGMLRESRLLAHLPKEMTQDIEQQLGSLILATDINRQNEFLTRLKAHLHN
KDLRLEDAQDRHFMLQIALKCADICNPCRIWEMSKQWSERVCEEFYRQGELEQKFELEIS
PLCNQQKDSIPSIQIGFMSYIVEPLFREWAHFTGNSTLSENMLGHLAHNKAQWKSLLPRQ
HRSRGSSGSGPDHDHAGQGTESEEQEGDSP
|
|
|
|---|
| Component 2 |
| Name: | High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | PDE7A | PDE7A_HUMAN | Phosphodiesterase 7 | Phosphodiesterase 7 (PDE7) | Phosphodiesterase 7A | Phosphodiesterase 7A (PDE7A1) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 55514.96 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q13946 |
|---|
| Residue: | 482 |
|---|
| Sequence: | MEVCYQLPVLPLDRPVPQHVLSRRGAISFSSSSALFGCPNPRQLSQRRGAISYDSSDQTA
LYIRMLGDVRVRSRAGFESERRGSHPYIDFRIFHSQSEIEVSVSARNIRRLLSFQRYLRS
SRFFRGTAVSNSLNILDDDYNGQAKCMLEKVGNWNFDIFLFDRLTNGNSLVSLTFHLFSL
HGLIEYFHLDMMKLRRFLVMIQEDYHSQNPYHNAVHAADVTQAMHCYLKEPKLANSVTPW
DILLSLIAAATHDLDHPGVNQPFLIKTNHYLATLYKNTSVLENHHWRSAVGLLRESGLFS
HLPLESRQQMETQIGALILATDISRQNEYLSLFRSHLDRGDLCLEDTRHRHLVLQMALKC
ADICNPCRTWELSKQWSEKVTEEFFHQGDIEKKYHLGVSPLCDRHTESIANIQIGFMTYL
VEPLFTEWARFSNTRLSQTMLGHVGLNKASWKGLQREQSSSEDTDAAFELNSQLLPQENR
LS
|
|
|
|---|
| BDBM23620 |
|---|
| n/a |
|---|
| Name | BDBM23620 |
|---|
| Synonyms: | 2-({6-[bis(2-hydroxyethyl)amino]-4,8-bis(piperidin-1-yl)pyrimido[5,4-d][1,3]diazin-2-yl}(2-hydroxyethyl)amino)ethan-1-ol | CHEMBL932 | Dipyridamine | Dipyridamole | Dipyudamine | MLS000028420 | Persantine | SMR000058382 | cid_3108 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H40N8O4 |
|---|
| Mol. Mass. | 504.6256 |
|---|
| SMILES | OCCN(CCO)c1nc(N2CCCCC2)c2nc(nc(N3CCCCC3)c2n1)N(CCO)CCO |
|---|
| Structure | 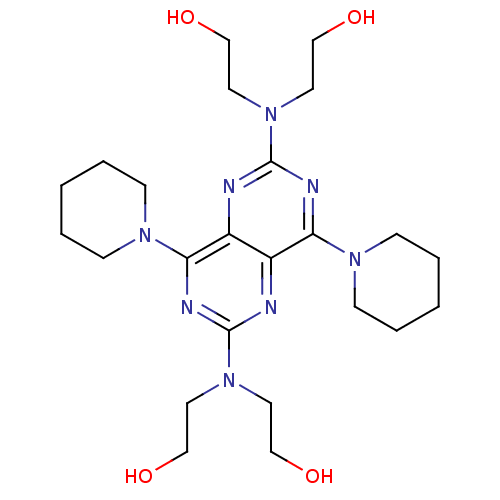
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Manallack, DT; Hughes, RA; Thompson, PE The next generation of phosphodiesterase inhibitors: structural clues to ligand and substrate selectivity of phosphodiesterases. J Med Chem48:3449-62 (2005) [PubMed] Article
Manallack, DT; Hughes, RA; Thompson, PE The next generation of phosphodiesterase inhibitors: structural clues to ligand and substrate selectivity of phosphodiesterases. J Med Chem48:3449-62 (2005) [PubMed] Article