

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cyclin-dependent kinase 6/G1/S-specific cyclin-D3 | ||
| Ligand | BDBM59227 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_473690 (CHEMBL936789) | ||
| IC50 | 282±n/a nM | ||
| Citation |  Fedorov, O; Marsden, B; Pogacic, V; Rellos, P; Müller, S; Bullock, AN; Schwaller, J; Sundström, M; Knapp, S A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A104:20523-8 (2007) [PubMed] Article Fedorov, O; Marsden, B; Pogacic, V; Rellos, P; Müller, S; Bullock, AN; Schwaller, J; Sundström, M; Knapp, S A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A104:20523-8 (2007) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cyclin-dependent kinase 6/G1/S-specific cyclin-D3 | |||
| Name: | Cyclin-dependent kinase 6/G1/S-specific cyclin-D3 | ||
| Synonyms: | CDK6/CycD3 | CDK6/D3 | CDK6/G1/S-specific cyclin D3 | CDK6/cyclin D3 | Cyclin-dependent kinase 6 | Cyclin-dependent kinase 6/G1/S-specific cyclin D3 | ||
| Type: | Protein | ||
| Mol. Mass.: | n/a | ||
| Description: | n/a | ||
| Components: | This complex has 2 components. | ||
| Component 1 | |||
| Name: | Cyclin-dependent kinase 6 | ||
| Synonyms: | CDK6 | CDK6_HUMAN | CDKN6 | Cell division protein kinase 6 | Cyclin-dependent kinase 6 (CDK 6) | Serine/threonine-protein kinase PLSTIRE | ||
| Type: | Enzyme Subunit | ||
| Mol. Mass.: | 36937.42 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q00534 | ||
| Residue: | 326 | ||
| Sequence: |
| ||
| Component 2 | |||
| Name: | G1/S-specific cyclin-D3 | ||
| Synonyms: | CCND3 | CCND3_HUMAN | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 32521.90 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P30281 | ||
| Residue: | 292 | ||
| Sequence: |
| ||
| BDBM59227 | |||
| n/a | |||
| Name | BDBM59227 | ||
| Synonyms: | Pyrazolopyrimidone analog, RGB-286147 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H22Cl2N4O3 | ||
| Mol. Mass. | 473.352 | ||
| SMILES | CC(C)c1nn(-c2c(Cl)cccc2Cl)c2nc(Cc3ccc(OCCO)cc3)[nH]c(=O)c12 |(-2.33,5.73,;-1.56,4.39,;-.02,4.39,;-2.33,3.06,;-1.43,1.81,;-2.33,.57,;-1.93,-.92,;-.44,-1.32,;.64,-.23,;-.05,-2.8,;-1.13,-3.89,;-2.62,-3.49,;-3.02,-2.01,;-4.51,-1.61,;-3.79,1.04,;-5.13,.27,;-6.46,1.04,;-7.8,.27,;-9.13,1.04,;-10.46,.27,;-11.8,1.04,;-11.8,2.58,;-13.13,3.35,;-14.46,2.58,;-15.8,3.35,;-17.13,2.58,;-10.46,3.35,;-9.13,2.58,;-6.46,2.58,;-5.13,3.35,;-5.13,4.89,;-3.79,2.58,)| | ||
| Structure | 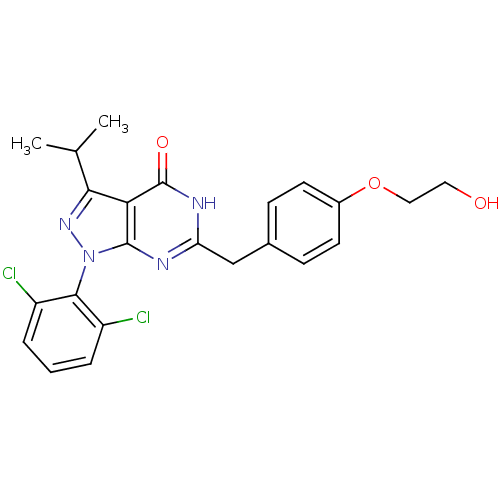
| ||