| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine racemase |
|---|
| Ligand | BDBM14673 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_594813 (CHEMBL1042552) |
|---|
| Ki | 71000±n/a nM |
|---|
| Citation |  Hoffman, HE; Jirásková, J; Cígler, P; Sanda, M; Schraml, J; Konvalinka, J Hydroxamic acids as a novel family of serine racemase inhibitors: mechanistic analysis reveals different modes of interaction with the pyridoxal-5'-phosphate cofactor. J Med Chem52:6032-41 (2009) [PubMed] Article Hoffman, HE; Jirásková, J; Cígler, P; Sanda, M; Schraml, J; Konvalinka, J Hydroxamic acids as a novel family of serine racemase inhibitors: mechanistic analysis reveals different modes of interaction with the pyridoxal-5'-phosphate cofactor. J Med Chem52:6032-41 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Serine racemase |
|---|
| Name: | Serine racemase |
|---|
| Synonyms: | D-serine ammonia-lyase | D-serine dehydratase | L-serine ammonia-lyase | L-serine dehydratase | SRR_MOUSE | Srr |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 36355.92 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_1441161 |
|---|
| Residue: | 339 |
|---|
| Sequence: | MCAQYCISFADVEKAHINIQDSIHLTPVLTSSILNQIAGRNLFFKCELFQKTGSFKIRGA
LNAIRGLIPDTPEEKPKAVVTHSSGNHGQALTYAAKLEGIPAYIVVPQTAPNCKKLAIQA
YGASIVYCDPSDESREKVTQRIMQETEGILVHPNQEPAVIAGQGTIALEVLNQVPLVDAL
VVPVGGGGMVAGIAITIKALKPSVKVYAAEPSNADDCYQSKLKGELTPNLHPPETIADGV
KSSIGLNTWPIIRDLVDDVFTVTEDEIKYATQLVWGRMKLLIEPTAGVALAAVLSQHFQT
VSPEVKNVCIVLSGGNVDLTSLNWVGQAERPAPYQTVSV
|
|
|
|---|
| BDBM14673 |
|---|
| n/a |
|---|
| Name | BDBM14673 |
|---|
| Synonyms: | Fragment 3 | Malonic Acid | propanedioic acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C3H4O4 |
|---|
| Mol. Mass. | 104.0615 |
|---|
| SMILES | OC(=O)CC(O)=O |
|---|
| Structure | 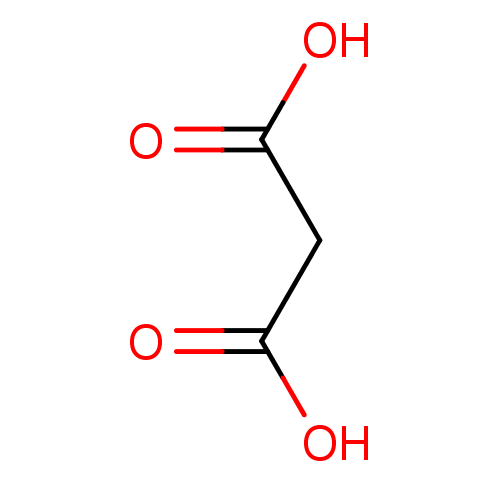
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hoffman, HE; Jirásková, J; Cígler, P; Sanda, M; Schraml, J; Konvalinka, J Hydroxamic acids as a novel family of serine racemase inhibitors: mechanistic analysis reveals different modes of interaction with the pyridoxal-5'-phosphate cofactor. J Med Chem52:6032-41 (2009) [PubMed] Article
Hoffman, HE; Jirásková, J; Cígler, P; Sanda, M; Schraml, J; Konvalinka, J Hydroxamic acids as a novel family of serine racemase inhibitors: mechanistic analysis reveals different modes of interaction with the pyridoxal-5'-phosphate cofactor. J Med Chem52:6032-41 (2009) [PubMed] Article