| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 3-phosphoinositide-dependent protein kinase 1 |
|---|
| Ligand | BDBM50321578 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_639726 (CHEMBL1175615) |
|---|
| IC50 | 2740±n/a nM |
|---|
| Citation |  Angiolini, M; Banfi, P; Casale, E; Casuscelli, F; Fiorelli, C; Saccardo, MB; Silvagni, M; Zuccotto, F Structure-based optimization of potent PDK1 inhibitors. Bioorg Med Chem Lett20:4095-9 (2010) [PubMed] Article Angiolini, M; Banfi, P; Casale, E; Casuscelli, F; Fiorelli, C; Saccardo, MB; Silvagni, M; Zuccotto, F Structure-based optimization of potent PDK1 inhibitors. Bioorg Med Chem Lett20:4095-9 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 3-phosphoinositide-dependent protein kinase 1 |
|---|
| Name: | 3-phosphoinositide-dependent protein kinase 1 |
|---|
| Synonyms: | 3-Phosphoinositide-Dependent Protein Kinase 1 (PDK1) | 3-phosphoinositide dependent protein kinase-1 | 3-phosphoinositide-dependent protein kinase 1 | 3-phosphoinositide-dependent protein kinase 1 (PDK) | 3-phosphoinositide-dependent protein kinase 1 (PDK-1) | 3-phosphoinositide-dependent protein kinase 1 (PDK1)(Δ1-50) | Isoform 2 of 3-phosphoinositide-dependent protein kinase 1 | PDK1 | PDPK1 | PDPK1_HUMAN | Phosphoinositide-dependent protein kinase 1 (PDK1) | Pyruvate dehydrogenase kinase isoenzyme 1 (PDK1) | hPDK1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 63157.65 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O15530 |
|---|
| Residue: | 556 |
|---|
| Sequence: | MARTTSQLYDAVPIQSSVVLCSCPSPSMVRTQTESSTPPGIPGGSRQGPAMDGTAAEPRP
GAGSLQHAQPPPQPRKKRPEDFKFGKILGEGSFSTVVLARELATSREYAIKILEKRHIIK
ENKVPYVTRERDVMSRLDHPFFVKLYFTFQDDEKLYFGLSYAKNGELLKYIRKIGSFDET
CTRFYTAEIVSALEYLHGKGIIHRDLKPENILLNEDMHIQITDFGTAKVLSPESKQARAN
SFVGTAQYVSPELLTEKSACKSSDLWALGCIIYQLVAGLPPFRAGNEYLIFQKIIKLEYD
FPEKFFPKARDLVEKLLVLDATKRLGCEEMEGYGPLKAHPFFESVTWENLHQQTPPKLTA
YLPAMSEDDEDCYGNYDNLLSQFGCMQVSSSSSSHSLSASDTGLPQRSGSNIEQYIHDLD
SNSFELDLQFSEDEKRLLLEKQAGGNPWHQFVENNLILKMGPVDKRKGLFARRRQLLLTE
GPHLYYVDPVNKVLKGEIPWSQELRPEAKNFKTFFVHTPNRTYYLMDPSGNAHKWCRKIQ
EVWRQRYQSHPDAAVQ
|
|
|
|---|
| BDBM50321578 |
|---|
| n/a |
|---|
| Name | BDBM50321578 |
|---|
| Synonyms: | 1-(2-amino-2-oxoethyl)-8-(phenylamino)-4,5-dihydro-1H-pyrazolo[4,3-h]quinazoline-3-carboxamide | CHEMBL1171948 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H17N7O2 |
|---|
| Mol. Mass. | 363.3733 |
|---|
| SMILES | NC(=O)Cn1nc(C(N)=O)c2CCc3cnc(Nc4ccccc4)nc3-c12 |
|---|
| Structure | 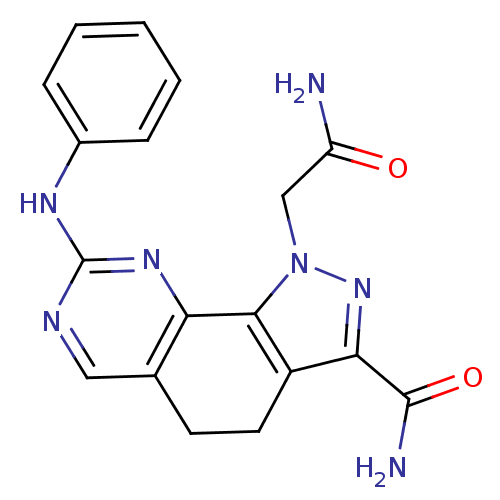
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Angiolini, M; Banfi, P; Casale, E; Casuscelli, F; Fiorelli, C; Saccardo, MB; Silvagni, M; Zuccotto, F Structure-based optimization of potent PDK1 inhibitors. Bioorg Med Chem Lett20:4095-9 (2010) [PubMed] Article
Angiolini, M; Banfi, P; Casale, E; Casuscelli, F; Fiorelli, C; Saccardo, MB; Silvagni, M; Zuccotto, F Structure-based optimization of potent PDK1 inhibitors. Bioorg Med Chem Lett20:4095-9 (2010) [PubMed] Article