| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Carbonic anhydrase 9 |
|---|
| Ligand | BDBM11625 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_643931 (CHEMBL1211830) |
|---|
| Ki | 38±n/a nM |
|---|
| Citation |  Avvaru, BS; Wagner, JM; Maresca, A; Scozzafava, A; Robbins, AH; Supuran, CT; McKenna, R Carbonic anhydrase inhibitors. The X-ray crystal structure of human isoform II in adduct with an adamantyl analogue of acetazolamide resides in a less utilized binding pocket than most hydrophobic inhibitors. Bioorg Med Chem Lett20:4376-81 (2010) [PubMed] Article Avvaru, BS; Wagner, JM; Maresca, A; Scozzafava, A; Robbins, AH; Supuran, CT; McKenna, R Carbonic anhydrase inhibitors. The X-ray crystal structure of human isoform II in adduct with an adamantyl analogue of acetazolamide resides in a less utilized binding pocket than most hydrophobic inhibitors. Bioorg Med Chem Lett20:4376-81 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Carbonic anhydrase 9 |
|---|
| Name: | Carbonic anhydrase 9 |
|---|
| Synonyms: | CA-IX | CA9 | CAH9_HUMAN | Carbonate dehydratase IX | Carbonic anhydrase 9 (CA IX) | Carbonic anhydrase 9 (CAIX) | Carbonic anhydrase 9 precursor | Carbonic anhydrase IX (CA IX) | Carbonic anhydrase IX (CAIX) | Carbonic anhydrases IX | Carbonic anhydrases; II & IX | G250 | MN | Membrane antigen MN | RCC-associated antigen G250 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 49669.03 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Catalytic domain of human cloned isozyme was used in the assay |
|---|
| Residue: | 459 |
|---|
| Sequence: | MAPLCPSPWLPLLIPAPAPGLTVQLLLSLLLLVPVHPQRLPRMQEDSPLGGGSSGEDDPL
GEEDLPSEEDSPREEDPPGEEDLPGEEDLPGEEDLPEVKPKSEEEGSLKLEDLPTVEAPG
DPQEPQNNAHRDKEGDDQSHWRYGGDPPWPRVSPACAGRFQSPVDIRPQLAAFCPALRPL
ELLGFQLPPLPELRLRNNGHSVQLTLPPGLEMALGPGREYRALQLHLHWGAAGRPGSEHT
VEGHRFPAEIHVVHLSTAFARVDEALGRPGGLAVLAAFLEEGPEENSAYEQLLSRLEEIA
EEGSETQVPGLDISALLPSDFSRYFQYEGSLTTPPCAQGVIWTVFNQTVMLSAKQLHTLS
DTLWGPGDSRLQLNFRATQPLNGRVIEASFPAGVDSSPRAAEPVQLNSCLAAGDILALVF
GLLFAVTSVAFLVQMRRQHRRGTKGGVSYRPAEVAETGA
|
|
|
|---|
| BDBM11625 |
|---|
| n/a |
|---|
| Name | BDBM11625 |
|---|
| Synonyms: | 2-N-(4-amino-3-chloro-5-fluorobenzene)-1,3,4-thiadiazole-2,5-disulfonamide | 5-{[(4-amino-3-chloro-5-fluorophenyl)sulfonyl]amino}-1,3,4-thiadiazole-2-sulfonamide | CHEMBL71611 | aminobenzolamide 18 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C8H7ClFN5O4S3 |
|---|
| Mol. Mass. | 387.819 |
|---|
| SMILES | Nc1c(F)cc(cc1Cl)S(=O)(=O)Nc1nnc(s1)S(N)(=O)=O |
|---|
| Structure | 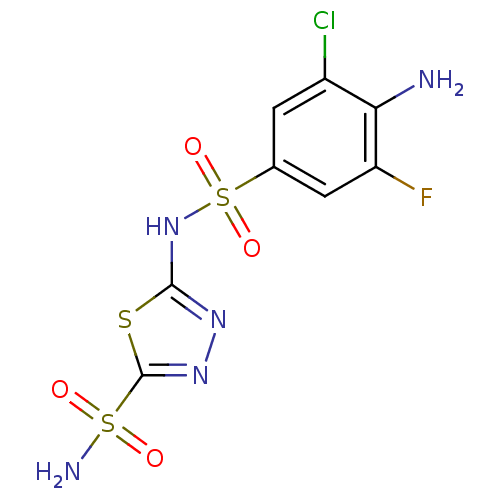
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Avvaru, BS; Wagner, JM; Maresca, A; Scozzafava, A; Robbins, AH; Supuran, CT; McKenna, R Carbonic anhydrase inhibitors. The X-ray crystal structure of human isoform II in adduct with an adamantyl analogue of acetazolamide resides in a less utilized binding pocket than most hydrophobic inhibitors. Bioorg Med Chem Lett20:4376-81 (2010) [PubMed] Article
Avvaru, BS; Wagner, JM; Maresca, A; Scozzafava, A; Robbins, AH; Supuran, CT; McKenna, R Carbonic anhydrase inhibitors. The X-ray crystal structure of human isoform II in adduct with an adamantyl analogue of acetazolamide resides in a less utilized binding pocket than most hydrophobic inhibitors. Bioorg Med Chem Lett20:4376-81 (2010) [PubMed] Article