| Reaction Details |
|---|
 | Report a problem with these data |
| Target | ATP-dependent translocase ABCB1 |
|---|
| Ligand | BDBM50295557 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_838281 (CHEMBL2076229) |
|---|
| Ki | 120000±n/a nM |
|---|
| Citation |  Ekins, S; Kim, RB; Leake, BF; Dantzig, AH; Schuetz, EG; Lan, LB; Yasuda, K; Shepard, RL; Winter, MA; Schuetz, JD; Wikel, JH; Wrighton, SA Three-dimensional quantitative structure-activity relationships of inhibitors of P-glycoprotein. Mol Pharmacol61:964-73 (2002) [PubMed] Ekins, S; Kim, RB; Leake, BF; Dantzig, AH; Schuetz, EG; Lan, LB; Yasuda, K; Shepard, RL; Winter, MA; Schuetz, JD; Wikel, JH; Wrighton, SA Three-dimensional quantitative structure-activity relationships of inhibitors of P-glycoprotein. Mol Pharmacol61:964-73 (2002) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| ATP-dependent translocase ABCB1 |
|---|
| Name: | ATP-dependent translocase ABCB1 |
|---|
| Synonyms: | ABCB1 | MDR1 | MDR1_HUMAN | Multidrug Resistance Transporter MDR 1 | Multidrug resistance protein 1 | Multidrug resistance protein 1/Multidrug resistance associated protein 1 | P-glycoprotein (P-gp) | P-glycoprotein 1 | PGY1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 141503.50 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P08183 |
|---|
| Residue: | 1280 |
|---|
| Sequence: | MDLEGDRNGGAKKKNFFKLNNKSEKDKKEKKPTVSVFSMFRYSNWLDKLYMVVGTLAAII
HGAGLPLMMLVFGEMTDIFANAGNLEDLMSNITNRSDINDTGFFMNLEEDMTRYAYYYSG
IGAGVLVAAYIQVSFWCLAAGRQIHKIRKQFFHAIMRQEIGWFDVHDVGELNTRLTDDVS
KINEGIGDKIGMFFQSMATFFTGFIVGFTRGWKLTLVILAISPVLGLSAAVWAKILSSFT
DKELLAYAKAGAVAEEVLAAIRTVIAFGGQKKELERYNKNLEEAKRIGIKKAITANISIG
AAFLLIYASYALAFWYGTTLVLSGEYSIGQVLTVFFSVLIGAFSVGQASPSIEAFANARG
AAYEIFKIIDNKPSIDSYSKSGHKPDNIKGNLEFRNVHFSYPSRKEVKILKGLNLKVQSG
QTVALVGNSGCGKSTTVQLMQRLYDPTEGMVSVDGQDIRTINVRFLREIIGVVSQEPVLF
ATTIAENIRYGRENVTMDEIEKAVKEANAYDFIMKLPHKFDTLVGERGAQLSGGQKQRIA
IARALVRNPKILLLDEATSALDTESEAVVQVALDKARKGRTTIVIAHRLSTVRNADVIAG
FDDGVIVEKGNHDELMKEKGIYFKLVTMQTAGNEVELENAADESKSEIDALEMSSNDSRS
SLIRKRSTRRSVRGSQAQDRKLSTKEALDESIPPVSFWRIMKLNLTEWPYFVVGVFCAII
NGGLQPAFAIIFSKIIGVFTRIDDPETKRQNSNLFSLLFLALGIISFITFFLQGFTFGKA
GEILTKRLRYMVFRSMLRQDVSWFDDPKNTTGALTTRLANDAAQVKGAIGSRLAVITQNI
ANLGTGIIISFIYGWQLTLLLLAIVPIIAIAGVVEMKMLSGQALKDKKELEGSGKIATEA
IENFRTVVSLTQEQKFEHMYAQSLQVPYRNSLRKAHIFGITFSFTQAMMYFSYAGCFRFG
AYLVAHKLMSFEDVLLVFSAVVFGAMAVGQVSSFAPDYAKAKISAAHIIMIIEKTPLIDS
YSTEGLMPNTLEGNVTFGEVVFNYPTRPDIPVLQGLSLEVKKGQTLALVGSSGCGKSTVV
QLLERFYDPLAGKVLLDGKEIKRLNVQWLRAHLGIVSQEPILFDCSIAENIAYGDNSRVV
SQEEIVRAAKEANIHAFIESLPNKYSTKVGDKGTQLSGGQKQRIAIARALVRQPHILLLD
EATSALDTESEKVVQEALDKAREGRTCIVIAHRLSTIQNADLIVVFQNGRVKEHGTHQQL
LAQKGIYFSMVSVQAGTKRQ
|
|
|
|---|
| BDBM50295557 |
|---|
| n/a |
|---|
| Name | BDBM50295557 |
|---|
| Synonyms: | 7-Methyl-4,6,6a,7,8,9,10,10a-octahydro-indolo[4,3-fg]quinoline-9-carboxylic acid ((2R,5S,10bS)-5-benzyl-10b-hydroxy-2-methyl-3,6-dioxo-octahydro-oxazolo[3,2-a]pyrrolo[2,1-c]pyrazin-2-yl)-amide | 7-Methyl-4,6,6a,7,8,9-hexahydro-indolo[4,3-fg]quinoline-9-carboxylic acid (5-benzyl-10b-hydroxy-2-methyl-3,6-dioxo-octahydro-oxazolo[3,2-a]pyrrolo[2,1-c]pyrazin-2-yl)-amide | CHEMBL1732 | DIHYDROERGOTAMINE | Dihydroer-gotamin |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C33H37N5O5 |
|---|
| Mol. Mass. | 583.6774 |
|---|
| SMILES | CN1C[C@@H](C[C@H]2[C@H]1Cc1c[nH]c3cccc2c13)C(=O)N[C@]1(C)O[C@@]2(O)[C@@H]3CCCN3C(=O)[C@H](Cc3ccccc3)N2C1=O |r| |
|---|
| Structure | 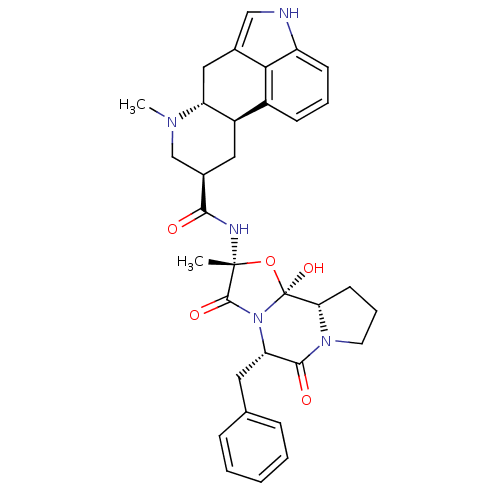
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ekins, S; Kim, RB; Leake, BF; Dantzig, AH; Schuetz, EG; Lan, LB; Yasuda, K; Shepard, RL; Winter, MA; Schuetz, JD; Wikel, JH; Wrighton, SA Three-dimensional quantitative structure-activity relationships of inhibitors of P-glycoprotein. Mol Pharmacol61:964-73 (2002) [PubMed]
Ekins, S; Kim, RB; Leake, BF; Dantzig, AH; Schuetz, EG; Lan, LB; Yasuda, K; Shepard, RL; Winter, MA; Schuetz, JD; Wikel, JH; Wrighton, SA Three-dimensional quantitative structure-activity relationships of inhibitors of P-glycoprotein. Mol Pharmacol61:964-73 (2002) [PubMed]