| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor alpha |
|---|
| Ligand | BDBM50099491 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_849356 (CHEMBL2149032) |
|---|
| EC50 | 0.6±n/a nM |
|---|
| Citation |  Baumgartner, L; Sosa, S; Atanasov, AG; Bodensieck, A; Fakhrudin, N; Bauer, J; Favero, GD; Ponti, C; Heiss, EH; Schwaiger, S; Ladurner, A; Widowitz, U; Loggia, RD; Rollinger, JM; Werz, O; Bauer, R; Dirsch, VM; Tubaro, A; Stuppner, H Lignan derivatives from Krameria lappacea roots inhibit acute inflammation in vivo and pro-inflammatory mediators in vitro. J Nat Prod74:1779-86 (2011) [PubMed] Article Baumgartner, L; Sosa, S; Atanasov, AG; Bodensieck, A; Fakhrudin, N; Bauer, J; Favero, GD; Ponti, C; Heiss, EH; Schwaiger, S; Ladurner, A; Widowitz, U; Loggia, RD; Rollinger, JM; Werz, O; Bauer, R; Dirsch, VM; Tubaro, A; Stuppner, H Lignan derivatives from Krameria lappacea roots inhibit acute inflammation in vivo and pro-inflammatory mediators in vitro. J Nat Prod74:1779-86 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor alpha |
|---|
| Name: | Peroxisome proliferator-activated receptor alpha |
|---|
| Synonyms: | NR1C1 | Nuclear receptor subfamily 1 group C member 1 | PPAR | PPAR alpha/gamma | PPAR-alpha | PPARA | PPARA_HUMAN | Peroxisome Proliferator-Activated Receptor alpha | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor alpha (PPAR alpha) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52222.08 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07869 |
|---|
| Residue: | 468 |
|---|
| Sequence: | MVDTESPLCPLSPLEAGDLESPLSEEFLQEMGNIQEISQSIGEDSSGSFGFTEYQYLGSC
PGSDGSVITDTLSPASSPSSVTYPVVPGSVDESPSGALNIECRICGDKASGYHYGVHACE
GCKGFFRRTIRLKLVYDKCDRSCKIQKKNRNKCQYCRFHKCLSVGMSHNAIRFGRMPRSE
KAKLKAEILTCEHDIEDSETADLKSLAKRIYEAYLKNFNMNKVKARVILSGKASNNPPFV
IHDMETLCMAEKTLVAKLVANGIQNKEAEVRIFHCCQCTSVETVTELTEFAKAIPGFANL
DLNDQVTLLKYGVYEAIFAMLSSVMNKDGMLVAYGNGFITREFLKSLRKPFCDIMEPKFD
FAMKFNALELDDSDISLFVAAIICCGDRPGLLNVGHIEKMQEGIVHVLRLHLQSNHPDDI
FLFPKLLQKMADLRQLVTEHAQLVQIIKKTESDAALHPLLQEIYRDMY
|
|
|
|---|
| BDBM50099491 |
|---|
| n/a |
|---|
| Name | BDBM50099491 |
|---|
| Synonyms: | 2-(4-(2-(3-cyclohexyl-1-(4-cyclohexylbutyl)ureido)ethyl)phenylthio)-2-methylpropanoic acid | 2-(4-{2-[3-Cyclohexyl-1-(4-cyclohexyl-butyl)-ureido]-ethyl}-phenylsulfanyl)-2-methyl-propionic acid | CHEMBL21241 | GW7647 | cid_3392731 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C29H46N2O3S |
|---|
| Mol. Mass. | 502.752 |
|---|
| SMILES | CC(C)(Sc1ccc(CCN(CCCCC2CCCCC2)C(=O)NC2CCCCC2)cc1)C(O)=O |
|---|
| Structure | 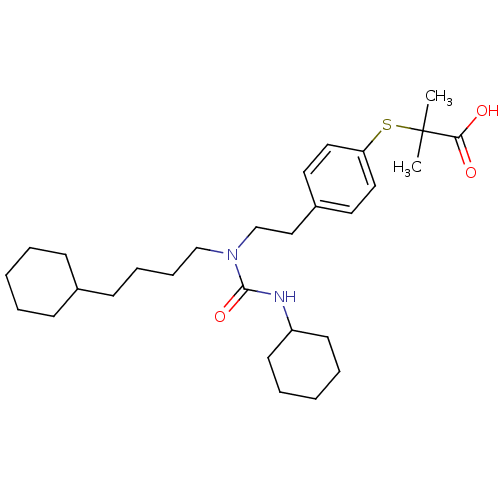
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Baumgartner, L; Sosa, S; Atanasov, AG; Bodensieck, A; Fakhrudin, N; Bauer, J; Favero, GD; Ponti, C; Heiss, EH; Schwaiger, S; Ladurner, A; Widowitz, U; Loggia, RD; Rollinger, JM; Werz, O; Bauer, R; Dirsch, VM; Tubaro, A; Stuppner, H Lignan derivatives from Krameria lappacea roots inhibit acute inflammation in vivo and pro-inflammatory mediators in vitro. J Nat Prod74:1779-86 (2011) [PubMed] Article
Baumgartner, L; Sosa, S; Atanasov, AG; Bodensieck, A; Fakhrudin, N; Bauer, J; Favero, GD; Ponti, C; Heiss, EH; Schwaiger, S; Ladurner, A; Widowitz, U; Loggia, RD; Rollinger, JM; Werz, O; Bauer, R; Dirsch, VM; Tubaro, A; Stuppner, H Lignan derivatives from Krameria lappacea roots inhibit acute inflammation in vivo and pro-inflammatory mediators in vitro. J Nat Prod74:1779-86 (2011) [PubMed] Article