| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Beta-secretase 1 |
|---|
| Ligand | BDBM50231937 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_872757 (CHEMBL2185582) |
|---|
| IC50 | 2.5±n/a nM |
|---|
| Citation |  Ghosh, AK; Venkateswara Rao, K; Yadav, ND; Anderson, DD; Gavande, N; Huang, X; Terzyan, S; Tang, J Structure-based design of highly selectiveß-secretase inhibitors: synthesis, biological evaluation, and protein-ligand X-ray crystal structure. J Med Chem55:9195-207 (2012) [PubMed] Article Ghosh, AK; Venkateswara Rao, K; Yadav, ND; Anderson, DD; Gavande, N; Huang, X; Terzyan, S; Tang, J Structure-based design of highly selectiveß-secretase inhibitors: synthesis, biological evaluation, and protein-ligand X-ray crystal structure. J Med Chem55:9195-207 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Beta-secretase 1 |
|---|
| Name: | Beta-secretase 1 |
|---|
| Synonyms: | ASP2 | Asp 2 | Aspartyl protease 2 | BACE | BACE1 | BACE1_HUMAN | Beta-secretase (BACE) | Beta-secretase 1 | Beta-secretase 1 (BACE 1) | Beta-secretase 1 (BACE-1) | Beta-secretase 1 (BACE1) | Beta-site APP cleaving enzyme 1 | Beta-site amyloid precursor protein cleaving enzyme 1 | KIAA1149 | Memapsin-2 | Membrane-associated aspartic protease 2 | beta-Secretase (BACE-1) | beta-Secretase (BACE1) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 55755.10 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P56817 |
|---|
| Residue: | 501 |
|---|
| Sequence: | MAQALPWLLLWMGAGVLPAHGTQHGIRLPLRSGLGGAPLGLRLPRETDEEPEEPGRRGSF
VEMVDNLRGKSGQGYYVEMTVGSPPQTLNILVDTGSSNFAVGAAPHPFLHRYYQRQLSST
YRDLRKGVYVPYTQGKWEGELGTDLVSIPHGPNVTVRANIAAITESDKFFINGSNWEGIL
GLAYAEIARPDDSLEPFFDSLVKQTHVPNLFSLQLCGAGFPLNQSEVLASVGGSMIIGGI
DHSLYTGSLWYTPIRREWYYEVIIVRVEINGQDLKMDCKEYNYDKSIVDSGTTNLRLPKK
VFEAAVKSIKAASSTEKFPDGFWLGEQLVCWQAGTTPWNIFPVISLYLMGEVTNQSFRIT
ILPQQYLRPVEDVATSQDDCYKFAISQSSTGTVMGAVIMEGFYVVFDRARKRIGFAVSAC
HVHDEFRTAAVEGPFVTLDMEDCGYNIPQTDESTLMTIAYVMAAICALFMLPLCLMVCQW
RCLRCLRQQHDDFADDISLLK
|
|
|
|---|
| BDBM50231937 |
|---|
| n/a |
|---|
| Name | BDBM50231937 |
|---|
| Synonyms: | CHEMBL403268 | N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | N1-((2S,3R)-4-(3-methoxybenzylamino)-3-hydroxy-1-phenylbutan-2-yl)-5-(N-methylmethan-3-ylsulfonamido)-N3-((R)-1-phenylethyl)isophthalamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C36H42N4O6S |
|---|
| Mol. Mass. | 658.807 |
|---|
| SMILES | COc1cccc(CNC[C@@H](O)[C@H](Cc2ccccc2)NC(=O)c2cc(cc(c2)C(=O)N[C@H](C)c2ccccc2)N(C)S(C)(=O)=O)c1 |
|---|
| Structure | 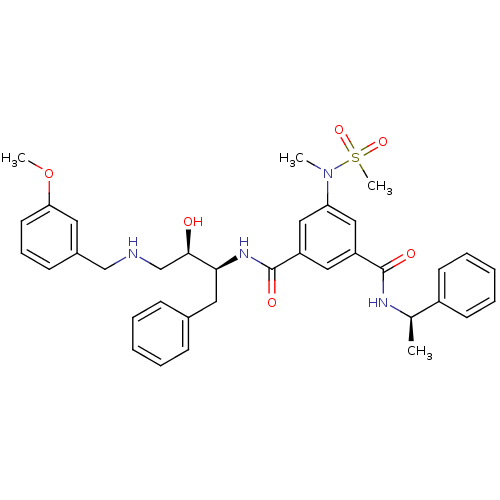
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ghosh, AK; Venkateswara Rao, K; Yadav, ND; Anderson, DD; Gavande, N; Huang, X; Terzyan, S; Tang, J Structure-based design of highly selectiveß-secretase inhibitors: synthesis, biological evaluation, and protein-ligand X-ray crystal structure. J Med Chem55:9195-207 (2012) [PubMed] Article
Ghosh, AK; Venkateswara Rao, K; Yadav, ND; Anderson, DD; Gavande, N; Huang, X; Terzyan, S; Tang, J Structure-based design of highly selectiveß-secretase inhibitors: synthesis, biological evaluation, and protein-ligand X-ray crystal structure. J Med Chem55:9195-207 (2012) [PubMed] Article